The genetic interaction map of the human solute carrier superfamily
- PMID: 40355755
- PMCID: PMC12130552
- DOI: 10.1038/s44320-025-00105-5
The genetic interaction map of the human solute carrier superfamily
Abstract
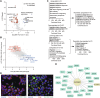
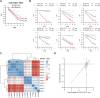
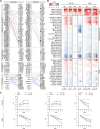
Solute carriers (SLCs), the largest superfamily of transporter proteins in humans with about 450 members, control the movement of molecules across membranes. A typical human cell expresses over 200 different SLCs, yet their collective influence on cell phenotypes is not well understood due to overlapping substrate specificities and expression patterns. To address this, we performed systematic pairwise gene double knockouts using CRISPR-Cas12a and -Cas9 in human colon carcinoma cells. A total of 1,088,605 guide combinations were used to interrogate 35,421 SLC-SLC and SLC-enzyme double knockout combinations across multiple growth conditions, uncovering 1236 genetic interactions with a growth phenotype. Further exploration of an interaction between the mitochondrial citrate/malate exchanger SLC25A1 and the zinc transporter SLC39A1 revealed an unexpected role for SLC39A1 in metabolic reprogramming and anti-apoptotic signaling. This full-scale genetic interaction map of human SLC transporters is the backbone for understanding the intricate functional network of SLCs in cellular systems and generates hypotheses for pharmacological target exploitation in cancer and other diseases. The results are available at https://re-solute.eu/resources/dashboards/genomics/ .
Keywords: CRISPR Screens; Functional Genomics; Genetic Interactions; Metabolism; Solute Carrier Transporters.
© 2025. The Author(s).
Conflict of interest statement
Disclosure and competing interests statement. GS-F is co-founder and owns shares of Solgate GmbH, an SLC-focused company. EG is the CSO of Solgate GmbH.
Figures







References
-
- Beijersbergen RL, Wessels LFA, Bernards R (2017) Synthetic lethality in cancer therapeutics. Annu Rev Cancer Biol 1:141–161
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

