Utilizing Nanopore direct RNA sequencing of blood from patients with sepsis for discovery of co- and post-transcriptional disease biomarkers
- PMID: 40355874
- PMCID: PMC12070577
- DOI: 10.1186/s12879-025-11078-z
Utilizing Nanopore direct RNA sequencing of blood from patients with sepsis for discovery of co- and post-transcriptional disease biomarkers
Abstract
Background: RNA sequencing of whole blood has been increasingly employed to find transcriptomic signatures of disease states. These studies traditionally utilize short-read sequencing of cDNA, missing important aspects of RNA expression such as differential isoform abundance and poly(A) tail length variation.
Methods: We used Oxford Nanopore Technologies sequencing to sequence native mRNA extracted from whole blood from 12 patients with definite bacterial and viral sepsis and compared with results from matching Illumina short-read cDNA sequencing data. Additionally, we explored poly(A) tail length variation, novel transcript identification, and differential transcript usage.
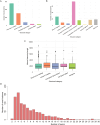
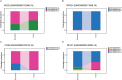
Results: The correlation of gene count data between Illumina cDNA- and Nanopore RNA-sequencing strongly depended on the choice of analysis pipeline; NanoCount for Nanopore and Kallisto for Illumina data yielded the highest mean Pearson's correlation of 0.927 at the gene level and 0.736 at the transcript isoform level. We identified 2 genes with differential polyadenylation, 9 genes with differential expression and 4 genes with differential transcript usage between bacterial and viral infection. Gene ontology gene set enrichment analysis of poly(A) tail length revealed enrichment of long tails in mRNA of genes involved in signaling and short tails in oxidoreductase molecular functions. Additionally, we detected 240 non-artifactual novel transcript isoforms.
Conclusions: Nanopore RNA- and Illumina cDNA-gene counts are strongly correlated, indicating that both platforms are suitable for discovery and validation of gene count biomarkers. Nanopore direct RNA-seq provides additional advantages by uncovering additional post- and co-transcriptional biomarkers, such as poly(A) tail length variation and transcript isoform usage.
Keywords: Differential transcript usage; Direct RNA-sequencing; Disease biomarkers; Long-read sequencing; Novel isoform detection; Oxford Nanopore Technologies; Polyadenylation.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: The Children’s Health Queensland Hospital and Health Service Human Research Ethics Committee; Queensland, Australia. approved the study on June 9, 2017 (HREC/17/QRCH/85). Written informed consent or delayed consent was obtained for all participants from their parents/carers. The study adhered to the WMA Declaration of Helsinki – Ethical Principles for Medical Research Involving Human Participants. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

