Testing Meningiomas With Methylation Arrays: Insights and Recommendations From a Large Single-Centre Study
- PMID: 40356449
- PMCID: PMC12070139
- DOI: 10.1111/nan.70018
Testing Meningiomas With Methylation Arrays: Insights and Recommendations From a Large Single-Centre Study
Abstract
Aims: Meningiomas are common primary CNS tumours, and their morphological diagnosis is usually straightforward. Their histological grading according to CNS WHO criteria alone provides limited information on recurrence risk. Risk stratification of meningiomas combining WHO grade, methylation class and copy number profile improves prediction of the risk of early recurrence. Because of the frequency of meningiomas in diagnostic practice, applying this prediction algorithm to all meningiomas is financially not viable in most healthcare systems.
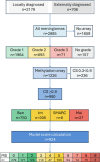
Methods: We analysed a retrospective dataset of over 1000 meningiomas from a single centre with methylation arrays to provide guidance on which meningiomas to prioritise for integrated molecular testing and to understand how WHO grades resolve into risk strata.
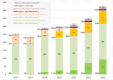
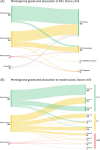
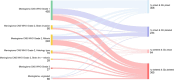
Results: Approximately 90% of CNS WHO Grade 1 meningiomas were allocated into the methylation class 'benign' and also into a low-risk group. Grade 2 meningiomas were allocated almost equally to either the low-risk (39%) or intermediate-risk groups (46%) but occasionally also to the high-risk group (15%). All grading criteria for CNS WHO Grade 2 meningiomas (brain invasion, mitotic count, cytoarchitectural atypia and histological type) showed a similar risk score distribution as the entire group. Grade 3 meningiomas were allocated to intermediate- (26%) or high-risk groups (74%).
Conclusion: Our data suggest that Grade 2 and 3 meningiomas should be prioritised for methylation profiling. A small proportion of Grade 1 meningiomas may also benefit from integrated molecular analysis, and further research is needed to explore if those histologically benign meningiomas with a predicted increased recurrence risk are associated with distinct demographic or histological characteristics.
Keywords: CNS WHO grading; meningioma; methylation array; model score; prognostication.
© 2025 The Author(s). Neuropathology and Applied Neurobiology published by John Wiley & Sons Ltd on behalf of British Neuropathological Society.
Conflict of interest statement
Felix Sahm is a co‐founder and shareholder of Heidelberg Epignostix GMBH, which licences the use of the methylation classifier. The other authors declare no conflicts of interest. Felix Sahm, Zane Jaunmuktane and Sebastian Brandner are members of the editorial board at Neuropathology and Applied Neurobiology.
Figures


References
-
- Perry A., Stafford S. L., Scheithauer B. W., Suman V. J., and Lohse C. M., “Meningioma Grading: An Analysis of Histologic Parameters,” American Journal of Surgical Pathology 21 (1997): 1455–1465. - PubMed
-
- Wang J. Z., Landry A. P., Raleigh D. R., et al., “Meningioma: International Consortium on Meningiomas (ICOM) Consensus Review on Scientific Advances & Treatment Paradigms for Clinicians, Researchers, and Patients,” Neuro‐Oncology 26, no. 10 (2024): 1742–1780, 10.1093/neuonc/noae082. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

