SuFEx-enabled high-throughput medicinal chemistry for developing potent tamoxifen analogs as Ebola virus entry inhibitors
- PMID: 40356906
- PMCID: PMC12066687
- DOI: 10.3389/fimmu.2025.1533037
SuFEx-enabled high-throughput medicinal chemistry for developing potent tamoxifen analogs as Ebola virus entry inhibitors
Abstract
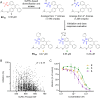
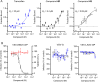
Ebola virus (EBOV) causes severe hemorrhagic fever with a high mortality rate in humans. In acute infection, an abnormal immune response results in excessive inflammatory cytokines and uncontrolled systemic inflammation that can result in organ damage and multi-organ failure. While vaccines and monoclonal antibody therapies are available, there is an urgent need for effective small-molecule antivirals against EBOV. Here, we report on the optimization of tamoxifen, an EBOV-glycoprotein (GP) binder that inhibits viral entry, using our Sulfur-Fluoride Exchange (SuFEx) click chemistry-based high-throughput medicinal chemistry (HTMC) strategy. Using a "Direct-to-Biology" approach, we generated a focused library of 2,496 tamoxifen analogs overnight and screened them in a cell-based pseudo-EBOV infection assay. The HTMC workflow enabled the development of a potent EBOV entry inhibitor with submicromolar EC50 cellular antiviral activity and more than 50-fold improvement in binding affinity against EBOV-GP compared to the parent compound. Our findings underscore the use of SuFEx-enabled HTMC for rapidly generating and assessing potential therapeutic candidates against viral and immune-mediated diseases in a cell-based assay.
Keywords: Ebola; SuFEx; direct-to-biology; drug discovery; small molecule antiviral drugs.
Copyright © 2025 Dada, Nagai, Agrawal, Wirchnianski, Wilson, Chandran and Kitamura.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

