Independent and temporally separated dynamics for RORγt and Foxp3 during Th17 differentiation
- PMID: 40356912
- PMCID: PMC12066577
- DOI: 10.3389/fimmu.2025.1462045
Independent and temporally separated dynamics for RORγt and Foxp3 during Th17 differentiation
Abstract
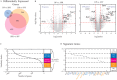
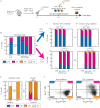
T helper 17 and Regulatory T cells (Th17 and Treg, respectively) are two well-described lymphocyte subsets with opposing actions. The divergent fates of Th17 and Treg cells are accounted for, at least in part, by molecular antagonism that occurs between their respective specific transcription factors, RORγt and Foxp3. An imbalance between Th17 and Treg cells may lead to tissue inflammation and is associated with certain types of autoimmunity. In order to understand the heterogeneity and dynamics of the differentiation process, we studied Th17/Treg cell differentiation of naïve cells in vitro, using RORγtGFPFoxp3RFP dual-reporter mouse. Flow cytometry revealed the consistent emergence of a population of double positive RORγt+Foxp3+ (DP) cells during the early stages of Th17 cell differentiation. These DP cells are closely related to RORγt+ single positive (SPR) cells in terms of global gene expression. Nevertheless, for some genes, DP cells share an expression pattern with Foxp3+ single positive (SPF) Treg cells, most importantly by reducing IL17 levels. Using time-lapse microscopy, we could delineate the expression dynamics of RORγt and Foxp3 at a clonal level. While the RORγt expression level elevates early during differentiation, Foxp3 rises later and is more stable upon environmental changes. These distinct expression profiles are independent of each other. During differentiation and proliferation, individual cells transit between SPR, DP, and SPF states. Nevertheless, the differentiation of sister cells within a clone progeny was highly correlated. We further demonstrated that sorted SPR and DP populations were not significantly affected by changes in their environment, suggesting that the correlated fate decision emerged at early time points before the first division. Overall, this study provides the first quantitative analysis of differentiation dynamics during the generation of DP RORγt+Foxp3+ cells. Characterizing these dynamics and the differentiation trajectory could provide a profound understanding and be used to better define the distinct fates of T cells, critical mediators of the immune response.
Keywords: T-cell differentiation; clonal analysis; double positive T-cells; micro-well array; systems immunology; time-lapse microscopy; transcription factor dynamics.
Copyright © 2025 Miller, Eizenberg-Magar, Reich-Zeliger, Rimer, Zaretsky, Reshef, Kopitman, Friedman and Antebi.
Conflict of interest statement
Author YEA is a scientific advisory board member and consultant at TeraCyte. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

