Unveiling p65 as the target of diphyllin in ameliorating metabolic dysfunction-associated steatotic liver disease via targeted protein degradation technology
- PMID: 40356997
- PMCID: PMC12066529
- DOI: 10.3389/fphar.2025.1567639
Unveiling p65 as the target of diphyllin in ameliorating metabolic dysfunction-associated steatotic liver disease via targeted protein degradation technology
Abstract
Introduction: Metabolic dysfunction-associated steatotic liver disease (MASLD), characterized by hepatic steatosis, inflammation and fibrosis, is becoming a global epidemic. However, the currently available effective clinical strategies remain limited.
Methods: We conducted the choline-deficient, L-amino acid-defined, high-fat diet (CDAHFD) induced MASH mouse model to explore the effects of diphyllin on MASLD mice. We employ the targeted protein degradation technology applied for the discovery of compound/protein-protein interaction to identify p65 as a potential target protein.
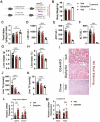
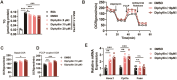
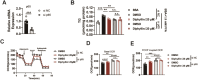
Results: We determine that diphyllin, a natural arylnaphthalene lignan lactone, is effective on MASLD, evidenced by the inhibition of hepatic lipid accumulation through promoting fatty acid oxidation in vivo and in vitro. To uncover the underlying mechanisms, we design and synthesis diphyllin-based protac and identify p65 as a potential target protein. Under p65 deficiency, the effects of diphyllin on lipid metabolism are blocked in vitro. As p65 as an antagonist of NRF2, diphyllin interacts with p65, leading to the induction of the NRF2 transcriptional activity and the enhancement of antioxidant capacity. When NFR2 is inhibited, the lowering effects of diphyllin on lipid is abolished.
Discussion: Our study presents diphyllin as a potential lead compound for MASLD therapy but also offers a novel approach for elucidating the mechanisms of action of natural products.
Keywords: NRF2; diphyllin; metabolic dysfunction-associated steatotic liver disease; p65; targeted protein degradation technology.
Copyright © 2025 Zhu, Zhang, Cui, Wang, Xu, Liu, Chen, Jiang, He, Peng and Li.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
LinkOut - more resources
Full Text Sources
Research Materials

