Advances in multi-trait genomic prediction approaches: classification, comparative analysis, and perspectives
- PMID: 40358423
- PMCID: PMC12070487
- DOI: 10.1093/bib/bbaf211
Advances in multi-trait genomic prediction approaches: classification, comparative analysis, and perspectives
Abstract
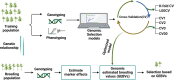
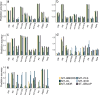
Traits in any organism are not independent, but show considerable integration, observed in a form of couplings and trade-offs. Therefore, improvement in one trait may affect other traits, often in undesired direction. To account for this problem, crop breeding increasingly relies on multi-trait genomic prediction (MT-GP) approaches that leverage the availability of genetic markers from different populations along with advances in high-throughput precision phenotyping. While significant progress has been made to jointly model multiple traits using a variety of statistical and machine learning approaches, there is no systematic comparison of advantages and shortcomings of the existing classes of MT-GP models. Here, we fill this knowledge gap by first classifying the existing MT-GP models and briefly summarizing their general principles, modeling assumptions, and potential limitations. We then perform an extensive comparative analysis with 10 traits measured in an Oryza sativa diversity panel using cross-validation scenarios relevant in breeding practice. Finally, we discuss directions that can enable the building of next generation MT-GP models in addressing pressing challenges in crop breeding.
Keywords: breeding; crop improvement; deep learning; genomic prediction; machine learning; multi-trait.
© The Author(s) 2025. Published by Oxford University Press.
Conflict of interest statement
All authors declare that they have no conflict of interest.
Figures
Similar articles
-
Genomic Selection in Plant Breeding: Methods, Models, and Perspectives.Trends Plant Sci. 2017 Nov;22(11):961-975. doi: 10.1016/j.tplants.2017.08.011. Epub 2017 Sep 28. Trends Plant Sci. 2017. PMID: 28965742 Review.
-
Integration of machine learning and genome-wide association study to explore the genomic prediction accuracy of agronomic trait in oats (Avena sativa L.).Plant Genome. 2025 Mar;18(1):e20549. doi: 10.1002/tpg2.20549. Plant Genome. 2025. PMID: 39780036 Free PMC article.
-
Machine learning approaches for crop improvement: Leveraging phenotypic and genotypic big data.J Plant Physiol. 2021 Feb;257:153354. doi: 10.1016/j.jplph.2020.153354. Epub 2020 Dec 29. J Plant Physiol. 2021. PMID: 33385619 Review.
-
GPOSYSH: Genomic Prediction of Oryza sativa Yield and Subpopulation Using Hybrid Methods.Recent Adv Food Nutr Agric. 2025;16(1):57-69. doi: 10.2174/012772574X281849240130120235. Recent Adv Food Nutr Agric. 2025. PMID: 38362690
-
mmGEBLUP: an advanced genomic prediction scheme for genetic improvement of complex traits in crops through integrative analysis of major genes, polygenes, and genotype-environment interactions.Brief Bioinform. 2024 Nov 22;26(1):bbaf058. doi: 10.1093/bib/bbaf058. Brief Bioinform. 2024. PMID: 39950744 Free PMC article.
Cited by
-
Elucidating Genotypic Variation in Quinoa via Multidimensional Agronomic, Physiological, and Biochemical Assessments.Plants (Basel). 2025 Jul 28;14(15):2332. doi: 10.3390/plants14152332. Plants (Basel). 2025. PMID: 40805681 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

