Relation equivariant graph neural networks to explore the mosaic-like tissue architecture of kidney diseases on spatially resolved transcriptomics
- PMID: 40358510
- PMCID: PMC12165735
- DOI: 10.1093/bioinformatics/btaf303
Relation equivariant graph neural networks to explore the mosaic-like tissue architecture of kidney diseases on spatially resolved transcriptomics
Abstract
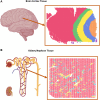
Motivation: Chronic kidney disease (CKD) and acute kidney injury (AKI) are prominent public health concerns affecting more than 15% of the global population. The ongoing development of spatially resolved transcriptomics (SRT) technologies presents a promising approach for discovering the spatial distribution patterns of gene expression within diseased tissues. However, existing computational tools are predominantly calibrated and designed on the ribbon-like structure of the brain cortex, presenting considerable computational obstacles in discerning highly heterogeneous mosaic-like tissue architectures in the kidney. Consequently, timely and cost-effective acquisition of annotation and interpretation in the kidney remains a challenge in exploring the cellular and morphological changes within renal tubules and their interstitial niches.
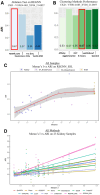
Results: We present an empowered graph deep learning framework, REGNN (Relation Equivariant Graph Neural Networks), designed for SRT data analyses on heterogeneous tissue structures. To increase expressive power in the SRT lattice using graph modeling, REGNN integrates equivariance to handle n-dimensional symmetries of the spatial area, while additionally leveraging Positional Encoding to strengthen relative spatial relations of the nodes uniformly distributed in the lattice. Given the limited availability of well-labeled spatial data, this framework implements both graph autoencoder and graph self-supervised learning strategies. On heterogeneous samples from different kidney conditions, REGNN outperforms existing computational tools in identifying tissue architectures within the 10× Visium platform. This framework offers a powerful graph deep learning tool for investigating tissues within highly heterogeneous expression patterns and paves the way to pinpoint underlying pathological mechanisms that contribute to the progression of complex diseases.
Availability and implementation: REGNN is publicly available at https://github.com/Mraina99/REGNN.
© The Author(s) 2025. Published by Oxford University Press.
Conflict of interest statement
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Figures



References
-
- 10xGenomics. ‘Human Breast Cancer,’ Single Cell Immune Profiling Dataset by Cell Ranger v3.1.0, 10x Genomics (2019, July 24).
-
- Abu-El-Haija S, Perozzi B, Kapoor A et al. MixHop: Higher-Order Graph Convolutional Architectures via Sparsified Neighborhood Mixing. In: International conference on machine learning (PMLR), 2019; 21–29.
-
- Allison SJ. Immune topology of the human kidney. Nat Rev Nephrol 2019;15:729. - PubMed
-
- Balcilar M, Héroux P, Gauzere B et al. Breaking the Limits of Message Passing Graph Neural Networks. In: Meila Marina and Zhang Tong (eds.), Proceedings of the 38th International Conference on Machine Learning (139; Proceedings of Machine Learning Research: PMLR), 2021; 599–608.
MeSH terms
Grants and funding
- U01DK114866/DK/NIDDK NIH HHS/United States
- UH3DK114861/DK/NIDDK NIH HHS/United States
- U01DK133766/DK/NIDDK NIH HHS/United States
- U01DK133081/DK/NIDDK NIH HHS/United States
- U01 DK114933/DK/NIDDK NIH HHS/United States
- U01 DK114908/DK/NIDDK NIH HHS/United States
- U01 DK133095/DK/NIDDK NIH HHS/United States
- U01 DK114907/DK/NIDDK NIH HHS/United States
- U54 AG075931/AG/NIA NIH HHS/United States
- U01DK133097/DK/NIDDK NIH HHS/United States
- U01DK133768/DK/NIDDK NIH HHS/United States
- U01DK114933/DK/NIDDK NIH HHS/United States
- U01 DK114923/DK/NIDDK NIH HHS/United States
- U24DK114886/DK/NIDDK NIH HHS/United States
- U01 DK133113/DK/NIDDK NIH HHS/United States
- Pelotonia Institute of Immuno-Oncology
- R35 GM126985/GM/NIGMS NIH HHS/United States
- R21HG012482/NH/NIH HHS/United States
- U01 DK114866/DK/NIDDK NIH HHS/United States
- R01 GM152585/GM/NIGMS NIH HHS/United States
- U01DK114908/DK/NIDDK NIH HHS/United States
- U01DK133092/DK/NIDDK NIH HHS/United States
- U01 DK133097/DK/NIDDK NIH HHS/United States
- UH3DK114937/DK/NIDDK NIH HHS/United States
- U01DK133095/DK/NIDDK NIH HHS/United States
- U01 DK133090/DK/NIDDK NIH HHS/United States
- UH3 DK114915/DK/NIDDK NIH HHS/United States
- U54AG075931/NH/NIH HHS/United States
- U01DK133091/DK/NIDDK NIH HHS/United States
- U01DK114920/DK/NIDDK NIH HHS/United States
- U01 DK133768/DK/NIDDK NIH HHS/United States
- UH3 DK114861/DK/NIDDK NIH HHS/United States
- U01 DK133092/DK/NIDDK NIH HHS/United States
- R21 HG012482/HG/NHGRI NIH HHS/United States
- UH3DK114926/DK/NIDDK NIH HHS/United States
- UH3 DK114937/DK/NIDDK NIH HHS/United States
- UH3DK114915/DK/NIDDK NIH HHS/United States
- U01DK114907/DK/NIDDK NIH HHS/United States
- U01 DK133081/DK/NIDDK NIH HHS/United States
- UH3 DK114926/DK/NIDDK NIH HHS/United States
- U01DK114923/DK/NIDDK NIH HHS/United States
- U01 DK133091/DK/NIDDK NIH HHS/United States
- U01 DK133093/DK/NIDDK NIH HHS/United States
- R35GM126985/NH/NIH HHS/United States
- U01 DK114920/DK/NIDDK NIH HHS/United States
- U01DK133093/DK/NIDDK NIH HHS/United States
- U24 DK114886/DK/NIDDK NIH HHS/United States
- U01DK133090/DK/NIDDK NIH HHS/United States
- U01DK133113/DK/NIDDK NIH HHS/United States
- U01 DK133766/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical

