Lessons from 5 Years of Routine Whole-Genome Sequencing for Epidemiologic Surveillance of Shiga Toxin-Producing Escherichia coli, France, 2018-2022
- PMID: 40359096
- PMCID: PMC12078545
- DOI: 10.3201/eid3113.241950
Lessons from 5 Years of Routine Whole-Genome Sequencing for Epidemiologic Surveillance of Shiga Toxin-Producing Escherichia coli, France, 2018-2022
Abstract
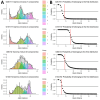
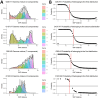
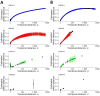
Whole-genome sequencing (WGS) is routine for surveillance of Shiga toxin-producing Escherichia coli human isolates in France. Protocols use EnteroBase hierarchical clustering at <5 allelic differences (HC5) as screening for cluster detection. We assessed current implementation after 5 years for 1,002 sequenced isolates. From genomic distances of serotypes O26:H11, O157:H7, O80:H2, and O103:H2, we determined statistical thresholds for cluster determination and compared those with HC5 clusters. Thresholds varied by serotype, 5-16 allelic distances and 15-20 single-nucleotide polymorphisms, showing limits of a single-threshold approach. We confirmed validity of HC5 screening for 3 serotypes because statistical thresholds had limited effect on isolate clustering (high sensitivity and specificity). For O80:H2, results suggest that HC5 is less reliable, and other approaches should be explored. Public health officials should regularly assess WGS used for Shiga toxin-producing E. coli surveillance to account for serotype and genomic evolution and to interpret WGS-linked isolates in light of epidemiologic data.
Keywords: France; STEC; Shiga toxin–producing Escherichia coli; bacteria; cluster detection; epidemiologic surveillance; hierarchical clustering; whole-genome sequencing.
Figures


References
-
- Bruyand M, Mariani-Kurkdjian P, Le Hello S, King LA, Van Cauteren D, Lefevre S, et al. ; Réseau français hospitalier de surveillance du SHU pédiatrique. Paediatric haemolytic uraemic syndrome related to Shiga toxin-producing Escherichia coli, an overview of 10 years of surveillance in France, 2007 to 2016. Euro Surveill. 2019;24:1800068. 10.2807/1560-7917.ES.2019.24.8.1800068 - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

