Fitness and transcriptional plasticity of human breast cancer single-cell-derived clones
- PMID: 40359107
- PMCID: PMC12117018
- DOI: 10.1016/j.celrep.2025.115699
Fitness and transcriptional plasticity of human breast cancer single-cell-derived clones
Abstract
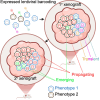
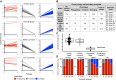
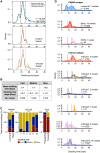
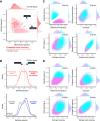
Clonal fitness and plasticity drive cancer heterogeneity. We used expressed lentiviral-based cellular barcodes combined with single-cell RNA sequencing to associate single-cell profiles with in vivo clonal growth. This generated a significant resource of growth measurements from over 20,000 single-cell-derived clones in 110 xenografts from 26 patient-derived breast cancer xenograft models. 167,375 single-cell RNA profiles were obtained from 5 models and revealed that rare propagating clones display a highly conserved model-specific differentiation program with reproducible regeneration of the entire transcriptomic landscape of the original xenograft. In 2 models of basal breast cancer, propagating clones demonstrated remarkable transcriptional plasticity at single-cell resolution. Dichotomous cell populations with different clonal growth properties, signaling pathways, and metabolic programs were characterized. By directly linking clonal growth with single-cell transcriptomes, these findings provide a profound understanding of clonal fitness and plasticity with implications for cancer biology and therapy.
Keywords: CP: Cancer; breast cancer; cancer stem cells; cellular barcoding; clonal heterogeneity; clonal tracking; patient-derived tumor xenografts; plasticity; single-cell sequencing.
Copyright © 2025 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests C.C. was in the past a recipient of research grants (administered by the University of Cambridge) from Genentech, Roche, AstraZeneca, and Servier.
Figures



References
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

