Alterations of ocular surface microbiome in glaucoma and its association with dry eye
- PMID: 40359128
- PMCID: PMC12075858
- DOI: 10.1099/jmm.0.002013
Alterations of ocular surface microbiome in glaucoma and its association with dry eye
Abstract
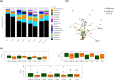
Introduction. Alterations in ocular surface microbiota (OSM) have been noted in both dry eye disease (DED) and glaucoma. However, the combined effects of these conditions on OSM have not been explored.Hypothesis. We hypothesized that patients with both glaucoma and dry eye would exhibit distinct changes in OSM composition and diversity compared to those with only glaucoma, only dry eye or healthy individuals.Aim. We employed amplicon sequencing to investigate OSM profiles in patients with glaucoma and/or dry eye disease.Methods. Swab samples from the conjunctiva of both eyes were collected from 28 glaucomatous patients [13 without dry eye syndrome (G-only) and 15 with dry eye syndrome (G-DED)], 13 DED patients without glaucoma (DED-only) and 31 age-matched healthy controls (HCs). After V3-V4 16S rRNA sequencing, MOTHUR tools and R language were used to elucidate and compare OSM composition and diversity between groups.Results. Our data revealed very diverse bacterial communities with 28 phyla and 785 genera. All the groups shared the three most abundant phyla, Actinobacteria (67.47%), Firmicutes (17.14%) and Proteobacteria (13.73%). Corynebacterium (54.75%), Staphylococcus (10.71%), Cutibacterium (8.77%) and Streptococcus (3.20%) were the most abundant genera. Only the G-DED group showed higher alpha diversity than the HC group (P<0.05). However, significant differences in beta diversity were observed between all three patient groups and the HC group. The Differential Expression for Sequencing 2 (DESeq2) analysis unveiled an increased presence of opportunistic bacteria across all pathological groups, with the G-DED group demonstrating the most pronounced alterations.Conclusions. Our findings confirm the predominance of Gram-positive bacteria in normal OSM and the rise of opportunistic Gram-negative bacteria in glaucoma and dry eye disease. This is the first study to characterize OSM in glaucoma patients with DED.
Keywords: conjunctiva; dry eye disease; dysbiosis; glaucoma; microbiome; preservatives.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures





References
-
- Tham YC, Li X, Wong TY, Quigley HA, Aung T, et al. Global prevalence of glaucoma and projections of glaucoma burden through 2040: a systematic review and meta-analysis. Ophthalmology. 2014;121:2081–2090. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

