Ribosome association inhibits stress-induced gene mRNA localization to stress granules
- PMID: 40360199
- PMCID: PMC12212004
- DOI: 10.1101/gad.352899.125
Ribosome association inhibits stress-induced gene mRNA localization to stress granules
Abstract
The integrated stress response (ISR) is critical for resilience to stress and is implicated in numerous diseases. During the ISR, translation is repressed, stress-induced genes are expressed, and mRNAs condense into stress granules. The relationship between stress granules and stress-induced gene expression is unclear. We measured endogenous stress-induced gene mRNA localization at the single-molecule level in the presence or absence of small molecule translation inhibitors. Reducing ribosome association increases the localization of stress-induced gene mRNAs to stress granules, whereas increasing ribosome association inhibits their localization to stress granules. The presence of upstream open reading frames (uORFs) in mRNA reporters reduces their localization to stress granules in a ribosome-dependent manner. Furthermore, a single initiating ribosome blocks stress granule formation and inhibits mRNA association with preformed stress granules. Thus, uORF-mediated ribosome association inhibits stress-induced gene mRNA localization to stress granules, suggesting a new role for uORFs in limiting RNA condensation.
Keywords: ATF4; GADD34; RNA localization; condensate; integrated stress response; ribosome; stress granules; stress-induced genes; translation; upstream open reading frame.
© 2025 Helton et al.; Published by Cold Spring Harbor Laboratory Press.
Conflict of interest statement
The authors declare no competing interests.
Figures
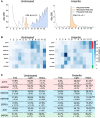

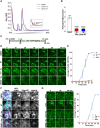
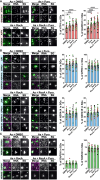
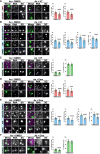


References
-
- Abadi M, Barham P, Chen J, Chen Z, Davis A, Dean J, Devin M, Ghemawat S, Irving G, Isard M, et al. 2016. TensorFlow: a system for large-scale machine learning. OSDI'16: Proceedings of the 12th USENIX Symposium on Operating Systems Design and Implementation, pp. 265–283. USENIX Association, San Francisco. https://www.usenix.org/system/files/conference/osdi16/osdi16-abadi.pdf
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
