Combination of a therapeutic cancer vaccine targeting the endogenous retroviral envelope protein ERVMER34-1 with immune-oncology agents facilitates expansion of neoepitope-specific T cells and promotes tumor control
- PMID: 40360436
- PMCID: PMC12163335
- DOI: 10.1136/jitc-2024-011378
Combination of a therapeutic cancer vaccine targeting the endogenous retroviral envelope protein ERVMER34-1 with immune-oncology agents facilitates expansion of neoepitope-specific T cells and promotes tumor control
Abstract
Background: Endogenous retroviruses (ERVs) are remnants of retrovirus germline infections that occurred over the course of evolution and constitute between 5% and 8% of the human genome. While ERVs tend to be epigenetically silenced in normal adult human tissues, they are often overexpressed in carcinomas and may represent novel immunotherapeutic targets. This study characterizes the ERV envelope protein ERVMER34-1 as a target for a therapeutic cancer vaccine.
Methods: The expression of ERVMER34-1 in multiple healthy adult and cancer tissues was assessed, as was its immunogenicity, to ascertain whether specific T cells could lyse human carcinoma cell lines expressing ERVMER34-1. Furthermore, the ability of a rationally designed ERVMER34-1-targeted therapeutic vaccine to induce tumor clearance in two murine carcinoma models expressing ERVMER34-1 was examined either as a monotherapy or in combination with anti-programmed cell death protein-1/programmed death-ligand 1 monoclonal antibody (mAb) or the interleukin-15 superagonist N-803.
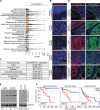
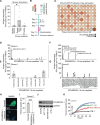
Results: The ERVMER34-1 protein was shown to be overexpressed in 232/376 of human carcinomas analyzed while being absent in most healthy adult tissues. High levels of ERVMER34-1 RNA expression associate with decreased survival in uveal melanoma, adenoid cystic, and head and neck carcinomas. ERVMER34-1-specific T cells were detected in peripheral blood mononuclear cells (PBMCs) of patients with cancer but not healthy donors following an overnight stimulation. However, reactive T cells are readily expanded from both healthy donor and patient with cancer PBMCs following a 7- day in vitro stimulation. Furthermore, ERVMER34-1-specific T cells selectively kill human carcinoma cell lines expressing ERVMER34-1. A novel, rationally designed, therapeutic cancer vaccine targeting ERVMER34-1 mediated tumor control in established syngeneic murine tumors expressing the full-length ERVMER34-1 protein. When combined with checkpoint blockade, the vaccine promoted expansion of neoepitope-reactive T cells whose function was further enhanced when combined with N-803. This expansion of neoepitope-reactive T cells was associated with tumor control.
Conclusions: This study reveals the potential of a vaccine that targets the retroviral envelope protein ERVMER34-1 and supports its continued development toward clinical testing as a new class of therapeutic cancer vaccine.
Keywords: Cytokine; Immune Checkpoint Inhibitor; Immunotherapy; Vaccine.
© Author(s) (or their employer(s)) 2025. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ Group.
Conflict of interest statement
Competing interests: The authors have no competing interests to disclose.
Figures



Similar articles
-
Combination Therapy Approaches to Enhance the Efficacy of ERV-Targeting Vaccines in Cancer.Cancer Immunol Res. 2025 Jun 4;13(6):792-803. doi: 10.1158/2326-6066.CIR-24-1192. Cancer Immunol Res. 2025. PMID: 40387511 Free PMC article. Review.
-
Immunomodulation to enhance the efficacy of an HPV therapeutic vaccine.J Immunother Cancer. 2020 Jun;8(1):e000612. doi: 10.1136/jitc-2020-000612. J Immunother Cancer. 2020. PMID: 32554612 Free PMC article.
-
New generation of DNA-based immunotherapy induces a potent immune response and increases the survival in different tumor models.J Immunother Cancer. 2021 Apr;9(4):e001243. doi: 10.1136/jitc-2020-001243. J Immunother Cancer. 2021. PMID: 33795383 Free PMC article.
-
Efficient Tumor Clearance and Diversified Immunity through Neoepitope Vaccines and Combinatorial Immunotherapy.Cancer Immunol Res. 2019 Aug;7(8):1359-1370. doi: 10.1158/2326-6066.CIR-18-0620. Epub 2019 Jul 10. Cancer Immunol Res. 2019. PMID: 31292145 Free PMC article.
-
Combination therapies utilizing neoepitope-targeted vaccines.Cancer Immunol Immunother. 2021 Apr;70(4):875-885. doi: 10.1007/s00262-020-02729-y. Epub 2020 Oct 8. Cancer Immunol Immunother. 2021. PMID: 33033852 Free PMC article. Review.
Cited by
-
The role of HERV envelope protein in ovarian cancer.Front Cell Dev Biol. 2025 Jul 31;13:1618542. doi: 10.3389/fcell.2025.1618542. eCollection 2025. Front Cell Dev Biol. 2025. PMID: 40823530 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
