TRIM23 mediates cGAS-induced autophagy in anti-HSV defense
- PMID: 40360474
- PMCID: PMC12075517
- DOI: 10.1038/s41467-025-59338-5
TRIM23 mediates cGAS-induced autophagy in anti-HSV defense
Erratum in
-
Publisher Correction: TRIM23 mediates cGAS-induced autophagy in anti-HSV defense.Nat Commun. 2025 Jul 3;16(1):6114. doi: 10.1038/s41467-025-61557-9. Nat Commun. 2025. PMID: 40610466 Free PMC article. No abstract available.
Abstract
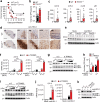
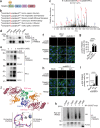
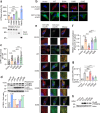
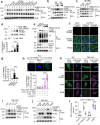
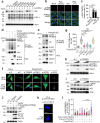
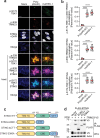
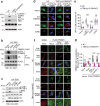
The cGAS-STING pathway, well-known to elicit interferon (IFN) responses, is also a key inducer of autophagy upon virus infection or other stimuli. Whereas the mediators for cGAS-induced IFN responses are well characterized, much less is known about how cGAS elicits autophagy. Here, we report that TRIM23, a unique TRIM protein harboring both ubiquitin E3 ligase and GTPase activity, is crucial for cGAS-STING-dependent antiviral autophagy. Genetic ablation of TRIM23 impairs autophagic control of HSV-1 infection. HSV-1 infection or cGAS-STING stimulation induces TBK1-mediated TRIM23 phosphorylation at S39, which triggers TRIM23 autoubiquitination and GTPase activity and ultimately elicits autophagy. Fibroblasts from a patient with herpes simplex encephalitis heterozygous for a dominant-negative, kinase-inactivating TBK1 mutation fail to activate autophagy by TRIM23 and cGAS-STING. Our results thus identify the cGAS-STING-TBK1-TRIM23 axis as a key autophagy defense pathway and may stimulate new therapeutic interventions for viral or inflammatory diseases.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

