Structural insight into the self-activation and G-protein coupling of P2Y2 receptor
- PMID: 40360475
- PMCID: PMC12075631
- DOI: 10.1038/s41421-025-00797-x
Structural insight into the self-activation and G-protein coupling of P2Y2 receptor
Abstract
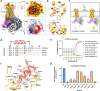
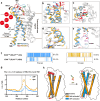
Purinergic P2Y2 receptor (P2Y2R) represents a typically extracellular ATP and UTP sensor for mediating purinergic signaling. Despite its importance as a pharmacological target, the molecular mechanisms underlying ligand recognition and G-protein coupling have remained elusive due to lack of structural information. In this study, we determined the cryo-electron microscopy (cryo-EM) structures of the apo P2Y2R in complex with Gq, ATP-bound P2Y2R in complex with Gq or Go, and UTP-bound P2Y4R in complex with Gq. These structures reveal the similarities and distinctions of ligand recognition within the P2Y receptor family. Furthermore, a comprehensive analysis of G-protein coupling reveals that P2Y2R exhibits promiscuity in coupling with both Gq and Go proteins. Combining molecular dynamics simulations and signaling assays, we elucidate the molecular mechanisms by which P2Y2R differentiates pathway-specific Gq or Go coupling through distinct structural components on the intracellular side. Strikingly, we identify a helix-like segment within the N-terminus that occupies the orthosteric ligand-binding pocket of P2Y2R, accounting for its self-activation. Taken together, these findings provide a molecular framework for understanding the activation mechanism of P2Y2R, encompassing ligand recognition, G-protein coupling, and a novel N-terminus-mediated self-activation mechanism.
© 2025. The Author(s).
Conflict of interest statement
Conflict of interest: The authors declare no competing interests.
Figures



Similar articles
-
Ligand binding and activation of UTP-activated G protein-coupled P2Y2 and P2Y4 receptors elucidated by mutagenesis, pharmacological and computational studies.Biochim Biophys Acta Gen Subj. 2020 Mar;1864(3):129501. doi: 10.1016/j.bbagen.2019.129501. Epub 2019 Dec 5. Biochim Biophys Acta Gen Subj. 2020. PMID: 31812541
-
P2Y2 nucleotide receptor activation enhances the aggregation and self-organization of dispersed salivary epithelial cells.Am J Physiol Cell Physiol. 2014 Jul 1;307(1):C83-96. doi: 10.1152/ajpcell.00380.2013. Epub 2014 Apr 23. Am J Physiol Cell Physiol. 2014. PMID: 24760984 Free PMC article.
-
Purinergic P2Y2 Receptor Control of Tissue Factor Transcription in Human Coronary Artery Endothelial Cells: NEW AP-1 TRANSCRIPTION FACTOR SITE AND NEGATIVE REGULATOR.J Biol Chem. 2016 Jan 22;291(4):1553-1563. doi: 10.1074/jbc.M115.681163. Epub 2015 Dec 2. J Biol Chem. 2016. PMID: 26631725 Free PMC article.
-
Therapeutic potential for P2Y2 receptor antagonism.Purinergic Signal. 2023 Jun;19(2):401-420. doi: 10.1007/s11302-022-09900-3. Epub 2022 Oct 11. Purinergic Signal. 2023. PMID: 36219327 Free PMC article. Review.
-
Tools and drugs for uracil nucleotide-activated P2Y receptors.Pharmacol Ther. 2018 Oct;190:24-80. doi: 10.1016/j.pharmthera.2018.04.002. Epub 2018 Apr 13. Pharmacol Ther. 2018. PMID: 29660366 Review.
References
-
- Burnstock, G. & Verkhratsky, A. Evolutionary origins of the purinergic signalling system. Acta Physiol.195, 415–447 (2009). - PubMed
-
- Rafehi, M. & Muller, C. E. Tools and drugs for uracil nucleotide-activated P2Y receptors. Pharm. Ther.190, 24–80 (2018). - PubMed
-
- Molliver, D. C., Cook, S. P., Carlsten, J. A., Wright, D. E. & McCleskey, E. W. ATP and UTP excite sensory neurons and induce CREB phosphorylation through the metabotropic receptor, P2Y2. Eur. J. Neurosci.16, 1850–1860 (2002). - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

