ATP functions as a pathogen-associated molecular pattern to activate the E3 ubiquitin ligase RNF213
- PMID: 40360510
- PMCID: PMC12075652
- DOI: 10.1038/s41467-025-59444-4
ATP functions as a pathogen-associated molecular pattern to activate the E3 ubiquitin ligase RNF213
Abstract
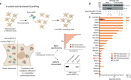
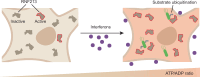
The giant E3 ubiquitin ligase RNF213 is a conserved component of mammalian cell-autonomous immunity, limiting the replication of bacteria, viruses and parasites. To understand how RNF213 reacts to these unrelated pathogens, we employ chemical and structural biology to find that ATP binding to its ATPases Associated with diverse cellular Activities (AAA) core activates its E3 function. We develop methodology for proteome-wide E3 activity profiling inside living cells, revealing that RNF213 undergoes a reversible switch in E3 activity in response to cellular ATP abundance. Interferon stimulation of macrophages raises intracellular ATP levels and primes RNF213 E3 activity, while glycolysis inhibition depletes ATP and downregulates E3 activity. These data imply that ATP bears hallmarks of a danger/pathogen associated molecular pattern, coordinating cell-autonomous defence. Furthermore, quantitative labelling of RNF213 with E3-activity probes enabled us to identify the catalytic cysteine required for substrate ubiquitination and obtain a cryo-EM structure of the RNF213-E2-ubiquitin conjugation enzyme transfer intermediate, illuminating an unannotated E2 docking site. Together, our data demonstrate that RNF213 represents a new class of ATP-dependent E3 enzyme, employing distinct catalytic and regulatory mechanisms adapted to its specialised role in the broad defence against intracellular pathogens.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: S.V. is an author of a patent relating to the ABP technology and is also Founder and shareholder of Outrun Therapeutics. The remaining authors declare no competing interests.
Figures




Similar articles
-
Moyamoya disease factor RNF213 is a giant E3 ligase with a dynein-like core and a distinct ubiquitin-transfer mechanism.Elife. 2020 Jun 23;9:e56185. doi: 10.7554/eLife.56185. Elife. 2020. PMID: 32573437 Free PMC article.
-
Interferon-Inducible E3 Ligase RNF213 Facilitates Host-Protective Linear and K63-Linked Ubiquitylation of Toxoplasma gondii Parasitophorous Vacuoles.mBio. 2022 Oct 26;13(5):e0188822. doi: 10.1128/mbio.01888-22. Epub 2022 Sep 26. mBio. 2022. PMID: 36154443 Free PMC article.
-
Moyamoya disease patient mutations in the RING domain of RNF213 reduce its ubiquitin ligase activity and enhance NFκB activation and apoptosis in an AAA+ domain-dependent manner.Biochem Biophys Res Commun. 2020 May 7;525(3):668-674. doi: 10.1016/j.bbrc.2020.02.024. Epub 2020 Mar 3. Biochem Biophys Res Commun. 2020. PMID: 32139119
-
The emerging role of E3 ubiquitin ligase RNF213 as an antimicrobial host determinant.Front Cell Infect Microbiol. 2023 Aug 15;13:1205355. doi: 10.3389/fcimb.2023.1205355. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 37655297 Free PMC article. Review.
-
Structural Diversity of Ubiquitin E3 Ligase.Molecules. 2021 Nov 4;26(21):6682. doi: 10.3390/molecules26216682. Molecules. 2021. PMID: 34771091 Free PMC article. Review.
References
-
- Kato, H. et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature441, 101–105 (2006). - PubMed
-
- Chamaillard, M. et al. An essential role for NOD1 in host recognition of bacterial peptidoglycan containing diaminopimelic acid. Nat. Immunol.4, 702–707 (2003). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

