SadB, a mediator of AmrZ proteolysis and biofilm development in Pseudomonas aeruginosa
- PMID: 40360526
- PMCID: PMC12075610
- DOI: 10.1038/s41522-025-00710-0
SadB, a mediator of AmrZ proteolysis and biofilm development in Pseudomonas aeruginosa
Abstract
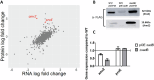
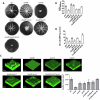
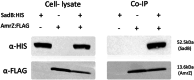
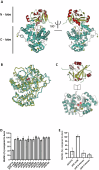
The ability of bacteria to commit to surface colonization and biofilm formation is a highly regulated process. In this study, we characterized the activity and structure of SadB, initially identified as a key regulator in the transition from reversible to irreversible surface attachment. Our results show that SadB acts as an adaptor protein that tightly regulates the master regulator AmrZ at the post-translational level. SadB directly binds to the C-terminal domain of AmrZ, leading to its rapid degradation, primarily by the Lon protease. Structural analysis suggests that SadB does not directly interact with small molecules upon signal transduction, differing from previous findings in Pseudomonas fluorescens. Instead, the SadB structure supports its role in mediating protein-protein interactions, establishing it as a major checkpoint for biofilm commitment.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

