Intragenic viral silencer element regulates HTLV-1 latency via RUNX complex recruitment
- PMID: 40360701
- PMCID: PMC12137137
- DOI: 10.1038/s41564-025-02006-7
Intragenic viral silencer element regulates HTLV-1 latency via RUNX complex recruitment
Abstract
Retroviruses integrate their genetic material into the host genome, enabling persistent infection. Human T cell leukaemia virus type 1 (HTLV-1) and human immunodeficiency virus type 1 (HIV-1) share similarities in genome structure and target cells, yet their infection dynamics differ drastically. While HIV-1 leads to high viral replication and immune system collapse, HTLV-1 establishes latency, promoting the survival of infected cells and, in some cases, leading to leukaemia. The mechanisms underlying this latency preference remain unclear. Here we analyse blood samples from people with HTLV-1 and identify an open chromatin region within the HTLV-1 provirus that functions as a transcriptional silencer and regulates transcriptional burst. The host transcription factor RUNX1 binds to this open chromatin region, repressing viral expression. Mutation of this silencer enhances HTLV-1 replication and immunogenicity, while its insertion into HIV-1 suppresses viral production. These findings reveal a strategy by which HTLV-1 ensures long-term persistence, offering potential insights into retroviral evolution and therapeutic targets.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures






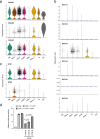

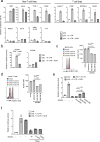





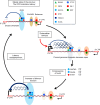
References
-
- Wolf, D. & Goff, S. P. TRIM28 mediates primer binding site-targeted silencing of murine leukemia virus in embryonic cells. Cell131, 46–57 (2007). - PubMed
-
- Matsui, T. et al. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature464, 927–931 (2010). - PubMed
-
- Proietti, F. A., Carneiro-Proietti, A. B., Catalan-Soares, B. C. & Murphy, E. L. Global epidemiology of HTLV-I infection and associated diseases. Oncogene24, 6058–6068 (2005). - PubMed
MeSH terms
Substances
Grants and funding
- JP20H03724/MEXT | Japan Society for the Promotion of Science (JSPS)
- JP21K08494/MEXT | Japan Society for the Promotion of Science (JSPS)
- JP23fk0410052/Japan Agency for Medical Research and Development (AMED)
- JP23wm0325068/Japan Agency for Medical Research and Development (AMED)
- JP23jm0210074/Japan Agency for Medical Research and Development (AMED)
LinkOut - more resources
Full Text Sources

