Morphological segmentation with tiling light sheet microscopy to quantitatively analyze the three-dimensional structures of spinal motoneurons
- PMID: 40360768
- PMCID: PMC12075063
- DOI: 10.1186/s13619-025-00231-3
Morphological segmentation with tiling light sheet microscopy to quantitatively analyze the three-dimensional structures of spinal motoneurons
Abstract
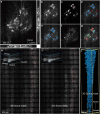
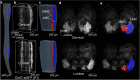
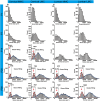
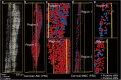
Spinal motoneurons control muscle fibers contraction and drive all motor behaviors in vertebrates. Although spinal motoneurons share the fundamental role of innervating muscle fibers, they exhibit remarkable diversity that reflects their specific identities. Defining the morphological changes during postnatal development is critical for elucidating this diversity. However, our understanding of the three-dimensional (3D) morphology of spinal motoneurons at these stages remains limited, largely due to the lack of high-throughput imaging tools. Using tiling light sheet microscopy combined with tissue clearing methods, we imaged motoneurons of the lateral and median motor column in the cervical and lumbar cord during postnatal development. By analyzing their soma size, we found that motoneurons innervating the upper limbs differentiate into two subpopulations with distinct soma size by postnatal day 14 (P14), while differentiation of motoneurons innervating the lower limbs is delayed. Furthermore, coupling adenovirus labeling with 3D volumetric reconstruction, we traced and measured the number and lengths of dendrites of flexor and extensor motoneurons in the lumbar cord, finding that the number of dendrites initially increases and subsequently declines as dendritic order rises. Together, these findings provide a quantitative analysis of the 3D morphological changes underlying spinal motoneuron diversity.
Keywords: 3D volumetric reconstruction; Dendritic arborization; Soma size; Spinal motoneurons; Tiling light sheet microscopy; Tissue clearing methods.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: The collection of mice tissues used in this study was approved by the Institutional Animal Care and Use Committee of the Westlake University (Approval No: 19 - 035-GL). Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures

References
-
- Ashrafi S, Lalancette-Hébert M, Friese A, Sigrist M, Arber S, Shneider NA, et al. Wnt7A identifies embryonic γ-motor neurons and reveals early postnatal dependence of γ-motor neurons on a muscle spindle-derived signal. J Neurosci. 2012;32(25):8725–31. 10.1523/jneurosci.1160-12.2012. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
