Early coordination of cell migration and cardiac fate determination during mammalian gastrulation
- PMID: 40360834
- PMCID: PMC12170898
- DOI: 10.1038/s44318-025-00441-0
Early coordination of cell migration and cardiac fate determination during mammalian gastrulation
Abstract
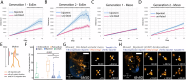
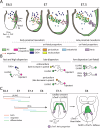
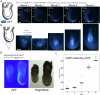
During gastrulation, mesodermal cells derived from distinct regions are destined to acquire specific cardiac fates after undergoing complex migratory movements. Here, we used light-sheet imaging of live mouse embryos between gastrulation and heart tube formation to track mesodermal cells and to reconstruct lineage trees and 3D migration paths for up to five cell divisions. We found independent progenitors emerging at specific times, contributing exclusively to left ventricle/atrioventricular canal (LV/AVC) or atrial myocytes. LV/AVC progenitors differentiated early to form the cardiac crescent, while atrial progenitors later generated the heart tube's Nr2f2+ inflow tract during morphogenesis. We also identified short-lived multipotent progenitors with broad potential, illustrating early developmental plasticity. Descendants of multipotent progenitors displayed greater dispersion and more diverse migratory trajectories within the anterior mesoderm than the progeny of uni-fated progenitors. Progenitors contributing to extraembryonic mesoderm (ExEm) exhibited the fastest and most dispersed migrations. In contrast, those giving rise to endocardial, LV/AVC, and pericardial cells showed a more gradual divergence, with late-stage behavioural shifts: endocardial cells increased in speed, while pericardial cells slowed down in comparison to LV/AVC cells. Together, these data reveal patterns of individual cell directionality and cardiac fate allocation within the seemingly unorganised migratory pattern of mesoderm cells.
Keywords: Cardiac Progenitors; Gastrulation; Heart Fields; Light Sheet Microscopy; Live imaging.
© 2025. The Author(s).
Conflict of interest statement
Disclosure and competing interests statement. The authors declare no competing interests.
Figures










Similar articles
-
Lineage contribution of the mesendoderm progenitors in the gastrulating mouse embryo.Dev Cell. 2025 Jul 21;60(14):1991-2006.e9. doi: 10.1016/j.devcel.2025.02.015. Epub 2025 Mar 24. Dev Cell. 2025. PMID: 40132585
-
Dynamic WT1 expression during gastrulation specifies peritoneal smooth muscle fate independently of mesothelial fate.Development. 2025 Jul 1;152(13):dev204332. doi: 10.1242/dev.204332. Epub 2025 Jul 9. Development. 2025. PMID: 40554754 Free PMC article.
-
Sertindole for schizophrenia.Cochrane Database Syst Rev. 2005 Jul 20;2005(3):CD001715. doi: 10.1002/14651858.CD001715.pub2. Cochrane Database Syst Rev. 2005. PMID: 16034864 Free PMC article.
-
Epiblast-derived CX3CR1+ progenitors generate cardiovascular cells during cardiogenesis.EMBO J. 2025 Aug;44(15):4331-4351. doi: 10.1038/s44318-025-00488-z. Epub 2025 Jun 23. EMBO J. 2025. PMID: 40551012 Free PMC article.
-
Drugs for preventing postoperative nausea and vomiting in adults after general anaesthesia: a network meta-analysis.Cochrane Database Syst Rev. 2020 Oct 19;10(10):CD012859. doi: 10.1002/14651858.CD012859.pub2. Cochrane Database Syst Rev. 2020. PMID: 33075160 Free PMC article.
References
-
- Aanhaanen WT, Brons JF, Dominguez JN, Rana MS, Norden J, Airik R, Wakker V, de Gier-de Vries C, Brown NA, Kispert A et al (2009) The Tbx2+ primary myocardium of the atrioventricular canal forms the atrioventricular node and the base of the left ventricle. Circ Res 104:1267–1274 - PubMed
-
- Bragantini J, Theodoro I, Zhao X, Huijben TA, Hirata-Miyasaki E, Vijay Kumar S, Balasubramanian A, Lao T, Agrawal R, Xiao S, Lammerding J, Mehta S, Falcão AX, Jacobo A, Lange M, Royer LA (2024) Ultrack: pushing the limits of cell tracking across biological scales. Preprint at https://www.biorxiv.org/content/10.1101/2024.09.02.610652v1 - DOI
-
- Chiba HSS (1978) Dynamic programming algorithm optimization for spoken word recognition. IEEE Trans Acoust Speech Signal Process 26:43–49
-
- Christiaen L, Davidson B, Kawashima T, Powell W, Nolla H, Vranizan K, Levine M (2008) The transcription/migration interface in heart precursors of Ciona intestinalis. Science 320:1349–1352 - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

