CHD1 loss reprograms SREBP2-driven cholesterol synthesis to fuel androgen-responsive growth and castration resistance in SPOP-mutated prostate tumors
- PMID: 40360905
- PMCID: PMC12276840
- DOI: 10.1038/s43018-025-00952-z
CHD1 loss reprograms SREBP2-driven cholesterol synthesis to fuel androgen-responsive growth and castration resistance in SPOP-mutated prostate tumors
Abstract
Despite undergoing castration, most individuals with prostate cancer (PCa) experience progression to castration-resistant PCa (CRPC), in which the androgen receptor (AR) remains an important driver. Concurrent genetic alterations in SPOP and CHD1 define a unique subtype of PCa, but their interactions in tumor progression and therapy response remain unclear. Here, we provide genetic evidence supporting that CHD1 loss accelerates disease progression and confers resistance to castration in males with SPOP-mutated PCa. By leveraging genetic engineering and multiomics, we uncovered a noncanonical function of CHD1 in lipid metabolism reprogramming via repressing the SREBP2 transcriptome. Loss of CHD1 induces cholesterol production, supplies intratumoral androgen biosynthesis and enhances AR activity, leading to castration resistance of SPOP-mutated PCa. Combining anti-androgen therapy with cholesterol-lowering drugs showed synergistic and durable activity against CRPC harboring CHD1 loss and SPOP mutations. These findings advance our understanding of an emerging PCa subtype and offer biomarker-driven combinatorial treatment strategies for men with CRPC.
© 2025. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Competing interests: D.E.F. has received research funding from GTx and has a familial relationship with March Biosciences, Biocity Biopharmaceuticals, Hummingbird Bioscience and Barricade Therapeutics. The other authors declare no competing interests.
Figures








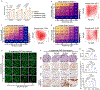

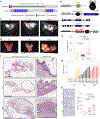
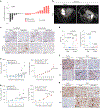




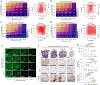

References
-
- Huggins C & Hodges CV Studies on prostatic cancer. I. The effect of castration, of estrogen and of androgen injection on serum phosphatases in metastatic carcinoma of the prostate. Cancer Res. 1, 293–297 (1941). - PubMed
-
- Stanbrough M et al. Increased expression of genes converting adrenal androgens to testosterone in androgen-independent prostate cancer. Cancer Res. 66, 2815–2825 (2006). - PubMed
MeSH terms
Substances
Grants and funding
- FP00016492/Prostate Cancer Foundation (PCF)
- R01CA247992/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- R01 CA269646/CA/NCI NIH HHS/United States
- R01CA244144/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- R01 CA181196/CA/NCI NIH HHS/United States
- R01 CA275990/CA/NCI NIH HHS/United States
- R01CA269646/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- PC230358/U.S. Department of Defense (United States Department of Defense)
- P30 CA016672/CA/NCI NIH HHS/United States
- R00 CA226360/CA/NCI NIH HHS/United States
- R01 CA275138/CA/NCI NIH HHS/United States
- R01CA181196/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- RP230072/Cancer Prevention and Research Institute of Texas (Cancer Prevention Research Institute of Texas)
- W81XWH-22-1-0686/U.S. Department of Defense (United States Department of Defense)
- CA275990/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- RR190021/Cancer Prevention and Research Institute of Texas (Cancer Prevention Research Institute of Texas)
- CA278889/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- R01 CA244144/CA/NCI NIH HHS/United States
- R01 CA247992/CA/NCI NIH HHS/United States
- CA275138/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- R01 CA278889/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Research Materials

