Generating diversity and securing completeness in algorithmic retrosynthesis
- PMID: 40361237
- PMCID: PMC12076909
- DOI: 10.1186/s13321-025-00981-x
Generating diversity and securing completeness in algorithmic retrosynthesis
Abstract
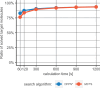
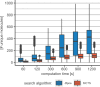
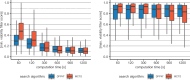
Chemical synthesis planning has considerably benefited from advances in the field of machine learning. Neural networks can reliably and accurately predict reactions leading to a given, possibly complex, molecule. In this work we focus on algorithms for assembling such predictions to a full synthesis plan that, starting from simple building blocks, produces a given target molecule, a procedure known as retrosynthesis. Objective functions for this task are hard to define and context-specific. In order to generate a diverse set of synthesis plans for chemists to select from, we capture the concept of diversity in a novel chemical diversity score (CDS). Our experiments show that our algorithm outperforms the algorithm predominantly employed in this domain, Monte-Carlo Tree Search, with respect to diversity in terms of our score as well as time efficiency. SCIENTIFIC CONTRIBUTION: We adapt Depth-First Proof-Number Search (DFPN) (Please refer to https://github.com/Bayer-Group/bayer-retrosynthesis-search for the accompanying source code.) and its variants, which have been applied to retrosynthesis before, to produce a set of solutions, with an explicit focus on diversity. We also make progress on understanding DFPN in terms of completeness, i.e., the ability to find a solution whenever there exists one. DFPN is known to be incomplete, for which we provide a much cleaner example, but we also show that it is complete when reinforced with a threshold-controlling routine from the literature.
Keywords: Chemical diversity score; Computer-Assisted Synthesis Planning (CASP); DFPN; Retrosynthesis.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing financial interest. Bayer AG was part of the MIT-led MLPDS consortium in the years 2018–2022.
Figures
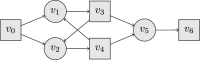





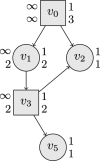

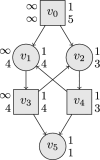

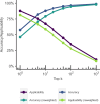
References
-
- Corey EJ, Long AK, Rubenstein SD (1985) Computer-assisted analysis in organic synthesis. Science 228(4698):408–418 - PubMed
-
- Corey EJ (1967) General methods for the construction of complex molecules. Pure Appl Chem 14(1):19–38
-
- Corey EJ, Wipke WT (1969) Computer-assisted design of complex organic syntheses: pathways for molecular synthesis can be devised with a computer and equipment for graphical communication. Science 166(3902):178–192 - PubMed
-
- Chen B, Li C, Dai H, Song L. Retro* (2020) Learning retrosynthetic planning with neural guided A* search. In International Conference on Machine Learning (ICML), 1608–1616
Grants and funding
LinkOut - more resources
Full Text Sources

