Identification of PECAM1 as a Prognostic Biomarker for Lung Adenocarcinoma
- PMID: 40361912
- PMCID: PMC12071333
- DOI: 10.3390/diagnostics15091094
Identification of PECAM1 as a Prognostic Biomarker for Lung Adenocarcinoma
Abstract
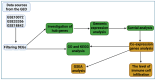
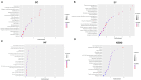
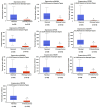
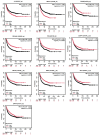
Background: Lung cancer continues to be one of the most fatal malignancies globally. Uncovering differentially expressed genes (DEGs) is crucial for advancing our understanding of tumor mechanisms and discovering new therapeutic targets. This study sought to identify key genes linked to prognosis and immune infiltration in lung cancer through the analysis of public gene expression datasets. Methods: We examined three microarray datasets from the Gene Expression Omnibus (GSE10072, GSE33356, and GSE18842) to detect DEGs between tumor and normal lung tissues. Functional enrichment was performed using Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses to interpret the biological relevance of these genes. Protein-protein interaction (PPI) networks were constructed via STRING and visualized using Cytoscape to screen for central hub genes. The prognostic implications of the hub genes were investigated using Kaplan-Meier Plotter and TIMER2.0 based on data from The Cancer Genome Atlas (TCGA). PECAM1 expression levels and its relationship with immune cell infiltration were further explored using UCSC Xena. Results: A total of 477 DEGs were consistently identified across all three datasets. Among the top 10 down-regulated hub genes, PECAM1 was significantly reduced in tumor tissues. Lower PECAM1 expression was positively associated with better first-progression survival (FPS) in lung cancer patients. This gene was particularly suppressed in lung adenocarcinoma (LUAD) and showed strong correlations with immune cell infiltration. Co-expression analysis revealed that genes linked to PECAM1 are involved in immune-related pathways. Conclusions: Our findings highlight PECAM1 as a potential prognostic biomarker in lung cancer, especially in LUAD. Its association with immune infiltration and patient survival supports its possible utility in early detection and as a candidate for immunotherapy development.
Keywords: DEGs; NSCLC; PECAM1; bioinformatics; immune infiltration; lung cancer.
Conflict of interest statement
The authors declare that they have no conflicts of interest.
Figures






References
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

