Comprehensive Characterization and Functional Analysis of the Lateral Organ Boundaries Domain Gene Family in Rice: Evolution, Expression, and Stress Response
- PMID: 40362188
- PMCID: PMC12071882
- DOI: 10.3390/ijms26093948
Comprehensive Characterization and Functional Analysis of the Lateral Organ Boundaries Domain Gene Family in Rice: Evolution, Expression, and Stress Response
Abstract
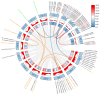
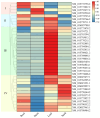
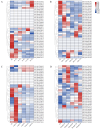
In this study, the LBD (Lateral Organ Boundaries Domain) gene family, a group of plant-specific transcription factors critical for plant growth and development as well as metabolic regulation, was comprehensively characterized in rice. We identified 36 LBD genes using multi-source genomic data and systematically classified them into Class I (31 genes) and Class II (5 genes). Analysis of their physicochemical properties revealed significant variations in amino acid length, molecular weight, isoelectric points, and hydropathicity. Motif analysis identified conserved LOB domains and other motifs potentially linked to functional diversity. Cis-acting element analysis indicated the involvement of these genes in various biological processes, including light response, hormone signaling, and stress response. Expression profiling demonstrated tissue-specific expression patterns, with several genes, such as XM_015770711.2, XM_015776632.2, and XM_015792766.2, showing relatively high expression in rice roots, implying their important role in root development. Transcriptome data further supported the involvement of specific genes in responses to phytohormones such as jasmonic acid (JA) and abscisic acid (ABA), as well as environmental stresses like cold and drought. Notably, XM_015770711.2, XM_015776632.2, and XM_015772758.2 may contribute to the regulation of rice environmental adaptability by mediating ABA and JA signaling pathways, respectively. In conclusion, this study identified members of the LBD gene family through the screening of two rice gene databases, and performed a comprehensive analysis of their physicochemical properties, evolutionary relationships, and expression profiles under various conditions. These findings provided valuable insights for further functional studies of LBD genes. Moreover, this study provides a foundation for targeting LBD genes to enhance stress resilience (e.g., drought/cold tolerance) and root architecture optimization. The LBD gene family possesses dual values in both stress resistance regulation and developmental optimization. The construction of its multidimensional functional map lays the theoretical and resource foundation for the precise design of high-yield and stress-resistant varieties.
Keywords: LBD gene family; gene expression; rice; stress response; system evolution.
Conflict of interest statement
The authors declare no potential conflict of interest with respect to the research, authorship, and/or publication of this article.
Figures

Similar articles
-
Genome-Wide Identification and Expression Analysis of Thionin Family in Rice (Oryza sativa) and Functional Characterization of OsTHION15 in Drought Stress and ABA Stress.Int J Mol Sci. 2025 Apr 7;26(7):3447. doi: 10.3390/ijms26073447. Int J Mol Sci. 2025. PMID: 40244412 Free PMC article.
-
Identification and Expression Analysis of CCCH Zinc Finger Family Genes in Oryza sativa.Genes (Basel). 2025 Apr 3;16(4):429. doi: 10.3390/genes16040429. Genes (Basel). 2025. PMID: 40282389 Free PMC article.
-
Expression dynamics indicate the role of Jasmonic acid biosynthesis pathway in regulating macronutrient (N, P and K+) deficiency tolerance in rice (Oryza sativa L.).Plant Cell Rep. 2021 Aug;40(8):1495-1512. doi: 10.1007/s00299-021-02721-5. Epub 2021 Jun 5. Plant Cell Rep. 2021. PMID: 34089089
-
Comprehensive characterization and gene expression patterns of LBD gene family in Gossypium.Planta. 2020 Mar 17;251(4):81. doi: 10.1007/s00425-020-03364-8. Planta. 2020. PMID: 32185507
-
JASMONATE ZIM-DOMAIN Family Proteins: Important Nodes in Jasmonic Acid-Abscisic Acid Crosstalk for Regulating Plant Response to Drought.Curr Protein Pept Sci. 2021 Dec 29;22(11):759-766. doi: 10.2174/1389203722666211018114443. Curr Protein Pept Sci. 2021. PMID: 34666642 Review.
References
-
- Iwakawa H., Ueno Y., Semiarti E., Onouchi H., Kojima S., Tsukaya H., Hasebe M., Soma T., Ikezaki M., Machida C., et al. The ASYMMETRIC LEAVES2 Gene of Arabidopsis thaliana, required for formation of a symmetric flat leaf lamina, encodes a member of a novel family of proteins characterized by cysteine repeats and a leucine zipper. Plant Cell Physiol. 2002;43:467–478. doi: 10.1093/pcp/pcf077. - DOI - PubMed
-
- Matsumura Y., Iwakawa H., Machida Y., Machida C. Characterization of genes in the ASYMMETRIC LEAVES2/LATERAL ORGAN BOUNDARIES (AS2/LOB) family in Arabidopsis thaliana, and functional and molecular comparisons between AS2 and other family members. Plant J. 2009;58:525–537. doi: 10.1111/j.1365-313X.2009.03797.x. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

