Probing Peptide Assembly and Interaction via High-Resolution Imaging Techniques: A Mini Review
- PMID: 40362238
- PMCID: PMC12071768
- DOI: 10.3390/ijms26093998
Probing Peptide Assembly and Interaction via High-Resolution Imaging Techniques: A Mini Review
Abstract
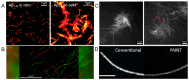
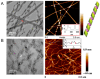
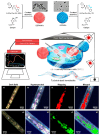
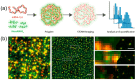
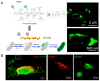
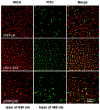
Peptide molecules, as fundamental structural units in biological systems, play pivotal roles in diverse biological processes and have garnered substantial attention in biomolecular self-assembly research. Their structural simplicity and high design flexibility make peptides key players in the development of novel biomaterials. High-resolution imaging techniques have provided profound insights into peptide assembly. Recently, the development of cutting-edge technologies, such as super-resolution microscopy (SRM) with unparalleled spatiotemporal resolution, has further advanced peptide assembly research. These advancements enable both the mechanistic exploration of peptide assembly pathways and the rational design of peptide-based functional materials. In this mini review, we systematically examine the structural diversity of peptide assemblies, including micelles, tubes, particles, fibers and hydrogel, as investigated by various high-resolution imaging techniques, with a focus on their assembly characterization and dynamic process. We also summarize the interaction networks of peptide assemblies with proteins, polymers and microbes, providing further insight into the interactions between peptide assemblies and other molecules. Furthermore, we emphasize the transformative role of high-resolution imaging techniques in addressing long-standing challenges in peptide nanotechnology. We anticipate that this review will accelerate the advancement of peptide assembly characterization, thereby fostering the creation of next-generation functional biomaterials.
Keywords: biomaterials; nanotechnology; peptide molecules; self-assembly; super-resolution microscopy.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

