Identification and Exploration of Pyroptosis-Related Genes in Macrophage Cells Reveal Necrotizing Enterocolitis Heterogeneity Through Single-Cell and Bulk-Sequencing
- PMID: 40362275
- PMCID: PMC12071306
- DOI: 10.3390/ijms26094036
Identification and Exploration of Pyroptosis-Related Genes in Macrophage Cells Reveal Necrotizing Enterocolitis Heterogeneity Through Single-Cell and Bulk-Sequencing
Abstract
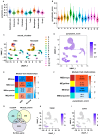
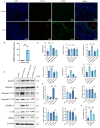
Necrotizing enterocolitis (NEC) is an acute intestine dysfunction intestinal disorder characterized by inflammation and cell death, including pyroptosis. Previous studies have implicated pyroptosis, particularly via NLRP3 inflammatory activation, and contribute to the development of NEC. However, the genetic and molecular mechanisms underlying pyroptosis in NEC pathogenesis and sequelae remain unclear. Our study aimed to identify the pyroptosis-related cell populations and genes and explore potential therapeutic targets. Single-cell RNA sequencing (scRNA-seq) data were analyzed to identify the cell populations related to NEC and pyroptosis. Weighted gene correlation network analysis (WGCNA) of bulk RNA-seq was performed to identify gene modules associate with pyroptosis. Cell-cell communication was employed to investigate intercellular signaling networks. Gene Set Enrichment Analysis (GSEA) was conducted to compare the pathways enriched in the high and low TREM1-expressing subgroups. Immunofluorescence staining was performed to detect the TREM1+CD163+ macrophages in the intestines. PCR and Western blot were performed to detect the expression of mRNA and proteins in the intestine tissues and cells. scRNA-seq analysis revealed increased macrophage abundance in NEC, with one macrophage cluster (cluster 4) exhibiting a markedly elevated pyroptosis score. WGCNA identified a gene module (MEbrown) that positively correlated with pyroptosis. Five genes (TREM1, TLN1, NOTCH2, MPZL1, and ADA) within this module were identified as potential diagnostic markers of pyroptosis. Furthermore, we identified a novel macrophage subpopulation, TREM1+CD163+, in NEC. Cell-cell communication analysis suggested that TREM1+CD163+ macrophages interact with other cells primarily through the NAMPT/ITGA5/ITGB1 and CCL3/CCR1 pathways. GSEA revealed a significant association between high TREM1 expression and pathways related to pyroptosis, cell proliferation, and inflammation. In vivo and in vitro experiments confirmed an increase in TREM1+CD163+ macrophages in NEC-affected intestines. TREM1 inhibition in THP-1 cells significantly reduced the expression of pro-inflammatory cytokines and pyroptosis-related genes and proteins. We identified the TREM1+CD163+ macrophage population that plays a crucial role in pyroptosis during NEC progression. Our findings elucidate the biological functions and molecular mechanisms of TREM1, demonstrating its upregulation in vivo and pro-pyroptosis effects in vitro. These insights advance our understanding of the role of pyroptosis in NEC pathogenesis and suggest TREM1 is a potential therapeutic target for NEC.
Keywords: TREM1; bioinformatics; macrophage; necrotizing enterocolitis; pyroptosis.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- Charoentong P., Finotello F., Angelova M., Mayer C., Efremova M., Rieder D., Hackl H., Trajanoski Z. Pan-cancer Immunogenomic Analyses Reveal Genotype-Immunophenotype Relationships and Predictors of Response to Checkpoint Blockade. Cell Rep. 2017;18:248–262. doi: 10.1016/j.celrep.2016.12.019. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

