Big Data-Driven Evolution of a Diagnostic Multiplex IgE-Test: Enhancing Accuracy and Efficacy in Allergy Diagnostics
- PMID: 40362486
- PMCID: PMC12072476
- DOI: 10.3390/ijms26094249
Big Data-Driven Evolution of a Diagnostic Multiplex IgE-Test: Enhancing Accuracy and Efficacy in Allergy Diagnostics
Abstract
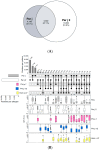
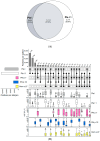
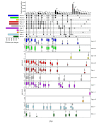
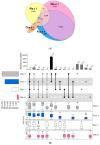
The ALEX2-test (MacroArray Diagnostics, Vienna, Austria) is a diagnostic multiplex IgE-test for the simultaneous detection of IgE to 178 allergens and 117 extracts, in addition to total IgE. Test results from more than 90 countries are stored on a GDPR-compliant cloud server for backup, customer support, and continuous postmarket surveillance. To improve the coverage of exposomes on a global scale and to further increase the sensitivity of the test, the allergen panel was updated from ALEX2 to ALEX3. By mid-2023, when ALEX3 was designed, almost 400,000 real-world ALEX2 test results were available. Analysing prevalences and average sIgE-levels of individual allergen preparations, coverage of extracts by components, and co-reactivity of members of the same allergen family provided a rationale for updating the array. In parallel, based on the scientific literature and clinical studies, new allergens were selected. On ALEX3, 218 allergens and 82 extracts will be represented, including 52 new allergens. Allergen preparations with low prevalence and clinical relevance, as well as redundant allergens and extracts, were discontinued. New allergens encompass, e.g., cyclophilins, alpha-gal, and additional markers from respiratory and food allergen sources. Using a large dataset of ALEX2 test results exemplifies the targeted, data-driven improvement of a diagnostic IgE-macroarray.
Keywords: ALEX; IgE-sensitisation; allergy; big-data; diagnosis; macroarray; microarray; molecular allergology; multiplex; test.
Conflict of interest statement
Peter Forstenlechner, Anna Ringauf, Raffaela Campana, Artan Salihu, Martina Aumayr, Irene Mittermann and Christian Lupinek are employees of MacroArray Diagnostics. The authors declare that this study received funding from MacroArray Diagnostics GmbH. The funder is the owner of the used data and of the described IgE-macroarray, i.e., ALEX. Purpose and methodology of the present study were as described in the present paper.
Figures




References
-
- Ansotegui I.J., Melioli G., Canonica G.W., Caraballo L., Villa E., Ebisawa M., Passalacqua G., Savi E., Ebo D., Gomez R.M., et al. IgE allergy diagnostics and other relevant tests in allergy, a World Allergy Organization position paper. World Allergy Organ. J. 2020;13:100080. doi: 10.1016/j.waojou.2019.100080. - DOI - PMC - PubMed
-
- Curin M., Reininger R., Swoboda I., Focke M., Valenta R., Spitzauer S. Skin prick test extracts for dog allergy diagnosis show considerable variations regarding the content of major and minor dog allergens. Int. Arch. Allergy Immunol. 2011;154:258–263. - PubMed
-
- Casset A., Mari A., Purohit A., Resch Y., Weghofer M., Ferrara R., Thomas W.R., Alessandri C., Chen K.W., de Blay F., et al. Varying allergen composition and content affects the in vivo allergenic activity of commercial Dermatophagoides pteronyssinus extracts. Int. Arch. Allergy Immunol. 2012;159:253–262. doi: 10.1159/000337654. - DOI - PMC - PubMed
-
- Glesner J., Filep S., Vailes L.D., Wunschmann S., Chapman M.D., Birrueta G., Frazier A., Jeong K.Y., Schal C., Bacharier L., et al. Allergen content in German cockroach extracts and sensitization profiles to a new expanded set of cockroach allergens determine in vitro extract potency for IgE reactivity. J. Allergy Clin. Immunol. 2019;143:1474–1481 e1478. - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

