Antibacterial Activity of the p53 Tumor Suppressor Protein-How Strong Is the Evidence?
- PMID: 40362653
- PMCID: PMC12072856
- DOI: 10.3390/ijms26094416
Antibacterial Activity of the p53 Tumor Suppressor Protein-How Strong Is the Evidence?
Abstract
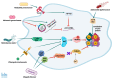
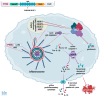
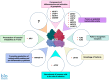
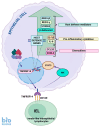
The p53 tumor suppressor is best known for controlling the cell cycle, apoptosis, DNA repair, and metabolism, but it also regulates immunity and is able to impede the live cycle of viruses. For this reason, these infectious agents encode proteins which inactivate p53. However, what is less known is that p53 can also be inactivated by human pathogenic bacteria. It is probably not due to collateral damage, but specific targeting, because p53 could interfere with their multiplication. The mechanisms of the antibacterial activity of p53 are poorly known. However, they can be inferred from the results of high-throughput studies, which have identified more than a thousand p53-activated genes. As it turns out, many of these genes code proteins which have proven or plausible antibacterial functions like the efficient detection of bacteria by pattern recognition receptors, the induction of pro-inflammatory pyroptosis, the recruitment of immune cells, direct bactericidal activity, and the presentation of bacterial metabolites to lymphocytes. Probably there are more antibacterial, p53-regulated functions which were overlooked because laboratory animals are kept in sterile conditions. In this review, we present the outlines of some intriguing antibacterial mechanisms of p53 which await further exploration. Definitely, this area of research deserves more attention, especially in light of the appearance of antibiotic-resistant bacterial strains.
Keywords: Helicobacter pylori; MDM2; actinomycin D; defensin; innate immunity; nutlin-3a; tuberculosis.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
References
-
- Łasut-Szyszka B., Gdowicz-Kłosok A., Małachowska B., Krześniak M., Będzińska A., Gawin M., Pietrowska M., Rusin M. Transcriptomic and proteomic study of cancer cell lines exposed to actinomycin D and nutlin-3a reveals numerous, novel candidates for p53-regulated genes. Chem. Biol. Interact. 2024;392:110946. doi: 10.1016/j.cbi.2024.110946. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

