iDDN: determining trans-omics network structure and rewiring with integrative differential dependency networks
- PMID: 40365110
- PMCID: PMC12070392
- DOI: 10.1093/bioadv/vbaf086
iDDN: determining trans-omics network structure and rewiring with integrative differential dependency networks
Abstract
Motivation: Mapping the gene networks that drive disease progression allows identifying molecules that rectify the network by normalizing pivotal regulatory elements. Upon mechanistic validation, these upstream normalizers represent attractive targets for developing therapeutic interventions to prevent the initiation or interrupt the pathways of disease progression. Differential network analysis aims to detect significant rewiring of regulatory network structures under different conditions. With few exceptions, most existing tools are limited to inferring differential networks from single-omics data that could be incomplete and prone to collapse when trans-omics multifactorial regulatory mechanisms are involved.
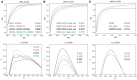
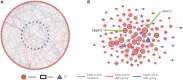
Results: We previously developed an efficient differential network analysis method-Differential Dependency Networks (DDN), that enables joint learning of common network structure and rewiring under different conditions. We now introduce the integrative DDN (iDDN) tool that extends this framework with biologically principled designs to make robust multi-omics differential network inferences. The comparative experimental evaluations on both realistic simulations and case studies show that iDDN can help biologists more accurately identify, in a study-specific and often unknown trans-omics regulatory circuitry, a network of differentially wired molecules potentially responsible for phenotypic transitions.
Availability and implementation: The Python package of iDDN is available at https://github.com/cbil-vt/iDDN. A user's guide is provided at https://iddn.readthedocs.io/.
© The Author(s) 2025. Published by Oxford University Press.
Conflict of interest statement
None declared.
Figures


Similar articles
-
DDN3.0: determining significant rewiring of biological network structure with differential dependency networks.Bioinformatics. 2024 Jun 3;40(6):btae376. doi: 10.1093/bioinformatics/btae376. Bioinformatics. 2024. PMID: 38902940 Free PMC article.
-
Assessing the comparative effects of interventions in COPD: a tutorial on network meta-analysis for clinicians.Respir Res. 2024 Dec 21;25(1):438. doi: 10.1186/s12931-024-03056-x. Respir Res. 2024. PMID: 39709425 Free PMC article. Review.
-
Molecular feature-based classification of retroperitoneal liposarcoma: a prospective cohort study.Elife. 2025 May 23;14:RP100887. doi: 10.7554/eLife.100887. Elife. 2025. PMID: 40407808 Free PMC article.
-
Surveillance for Violent Deaths - National Violent Death Reporting System, 50 States, the District of Columbia, and Puerto Rico, 2022.MMWR Surveill Summ. 2025 Jun 12;74(5):1-42. doi: 10.15585/mmwr.ss7405a1. MMWR Surveill Summ. 2025. PMID: 40493548 Free PMC article.
-
Pharmacological and electronic cigarette interventions for smoking cessation in adults: component network meta-analyses.Cochrane Database Syst Rev. 2023 Sep 12;9(9):CD015226. doi: 10.1002/14651858.CD015226.pub2. Cochrane Database Syst Rev. 2023. PMID: 37696529 Free PMC article.
References
-
- 10x Genomcis. PBMC from a healthy donor—granulocytes removed through cell sorting (10k). 2010. https://support.10xgenomics.com/single-cell-multiome-atac-gex/datasets/1...
-
- Cui H, Wang C, Maan H et al. scGPT: toward building a foundation model for single-cell multi-omics using generative AI. Nat Methods 2024;21:1470–80. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

