Investigating post-infection anxiety- and depression-like behaviors in a SARS-CoV-2 mouse model
- PMID: 40365287
- PMCID: PMC12068287
- DOI: 10.7150/thno.102752
Investigating post-infection anxiety- and depression-like behaviors in a SARS-CoV-2 mouse model
Abstract
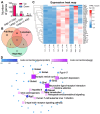
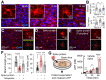
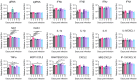
Rationale: The COVID-19 pandemic, driven by SARS-CoV-2, has resulted in a wide range of neuropsychiatric symptoms associated with post-acute sequelae (PASC). However, the mechanisms by which SARS-CoV-2 impacts the brain and leads to persistent behavioral changes remain poorly understood. We hypothesize that SARS-CoV-2 exposure induces neuroinflammation and microglial activation, leading to anxiety- and depression-like behaviors in mice. Methods: We established a SARS-CoV-2 mouse model using the virulent SARS2-N501YMA30 strain to investigate its impact on the central nervous system (CNS). We assessed neuroinvasion via immunostaining of dsRNA and markers for neuronal, astrocyte, and microglia in brain slices. Behavioral changes were evaluated at 2 weeks, 2 months, and 4 months post-infection. Molecular and cellular analyses included bulk RNA-seq, Golgi-Cox staining, field excitatory postsynaptic potential (fEPSP) recordings, immunofluorescence, and quantitative real-time PCR (qRT-PCR) to assess gene expression, neuronal morphology, and microglial activation in the brain. Results: We demonstrated that intranasal inoculation of SARS2-N501YMA30 results in viral dissemination to multiple brain regions, including the amygdala and the prefrontal cortex (PFC). Behavioral assays indicated a marked elevation in anxiety- and depression-like behaviors post-infection. A comparative analysis of RNA expression profiles disclosed alterations in the post-infected brains. Additionally, we observed dendritic spine remodeling on neurons within the amygdala after infection. Infection with SARS2-N501YMA30 was associated with microglial activation and a subsequent increase in microglia-dependent neuronal activity in the amygdala. Transcriptomic analysis of infected brains revealed the upregulation of inflammatory and cytokine-related pathways, implicating neuroinflammation in the pathogenesis of neuronal hyperactivity and behavioral abnormality. Conclusion: Our findings provide evidence that SARS-CoV-2 neuroinvasion plays a critical role in the development of lasting behavioral sequelae observed in PASC. These data provide critical insights into the neurological consequences of SARS-CoV-2 infection and underscore microglia as a potential therapeutic target for ameliorating virus-induced neurobehavioral abnormalities.
Keywords: Amygdala; Anxiety- and depression-like behaviors.; Microglia; Post-acute sequelae of COVID-19; SARS-CoV-2.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures



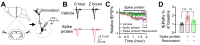
Similar articles
-
SARS-CoV-2 neurotropism-induced anxiety/depression-like behaviors require Microglia activation.bioRxiv [Preprint]. 2024 Jun 10:2023.10.02.560570. doi: 10.1101/2023.10.02.560570. bioRxiv. 2024. PMID: 37873397 Free PMC article. Preprint.
-
Targeting P2X7 mitigates neurobehavioural alterations in a mouse model of post-acute sequelae of SARS-CoV-2 infection.Neuropharmacology. 2025 Nov 1;278:110566. doi: 10.1016/j.neuropharm.2025.110566. Epub 2025 Jun 16. Neuropharmacology. 2025. PMID: 40532786 Review.
-
Stress-Induced Neuronal Colony Stimulating Factor 1 Provokes Microglia-Mediated Neuronal Remodeling and Depressive-like Behavior.Biol Psychiatry. 2018 Jan 1;83(1):38-49. doi: 10.1016/j.biopsych.2017.05.026. Epub 2017 Jun 12. Biol Psychiatry. 2018. PMID: 28697890 Free PMC article.
-
CB2R activation ameliorates late adolescent chronic alcohol exposure-induced anxiety-like behaviors during withdrawal by preventing morphological changes and suppressing NLRP3 inflammasome activation in prefrontal cortex microglia in mice.Brain Behav Immun. 2023 May;110:60-79. doi: 10.1016/j.bbi.2023.02.001. Epub 2023 Feb 6. Brain Behav Immun. 2023. PMID: 36754245
-
Pathogenesis Underlying Neurological Manifestations of Long COVID Syndrome and Potential Therapeutics.Cells. 2023 Mar 6;12(5):816. doi: 10.3390/cells12050816. Cells. 2023. PMID: 36899952 Free PMC article. Review.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

