Genome analysis reveals diverse novel psychrotolerant Mucilaginibacter species in Arctic tundra soils
- PMID: 40365603
- PMCID: PMC12074574
- DOI: 10.1093/ismeco/ycaf071
Genome analysis reveals diverse novel psychrotolerant Mucilaginibacter species in Arctic tundra soils
Abstract
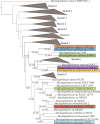
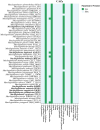
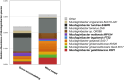
As Arctic soil ecosystems warm due to climate change, enhanced microbial activity is projected to increase the rate of soil organic matter degradation. Delineating the diversity and activity of Arctic tundra microbial communities active in decomposition is thus of keen interest. Here, we describe novel cold-adapted bacteria in the genus Mucilaginibacter (Bacteroidota) isolated from Artic tundra soils in Finland. These isolates are aerobic chemoorganotrophs and appear well adapted to the low-temperature environment, where they are also exposed to desiccation and a wide regime of annual temperature variation. Initial 16S ribosomal RNA (rRNA)-based phylogenetic analysis suggested that five isolated strains represent new species of the genus Mucilaginibacter, confirmed by whole genome-based phylogenomic and average nucleotide identity. Five novel species are described: Mucilaginibacter geliditolerans sp. nov., Mucilaginibacter tundrae sp. nov., Mucilaginibacter empetricola sp. nov., Mucilaginibacter saanensis sp. nov., and Mucilaginibacter cryoferens sp. nov. Genome and phenotype analysis showed their potential in complex carbon degradation, nitrogen assimilation, polyphenol degradation, and adaptation to their tundra heath habitat. A pangenome analysis of the newly identified species alongside known members of the Mucilaginibacter genus sourced from various environments revealed the distinctive characteristics of the tundra strains. These strains possess unique genes related to energy production, nitrogen uptake, adaptation, and the synthesis of secondary metabolites that aid in their growth, potentially accounting for their prevalence in tundra soil. By uncovering novel species and strains within the Mucilaginibacter, we enhance our understanding of this genus and elucidate how environmental fluctuations shape the microbial functionality and interactions in Arctic tundra ecosystems.
Keywords: Mucilaginibacter; cold-adapted; novel species; tundra soil isolates.
© The Author(s) 2025. Published by Oxford University Press on behalf of the International Society for Microbial Ecology.
Conflict of interest statement
None declared.
Figures



Similar articles
-
Genome analysis and description of Tunturibacter gen. nov. expands the diversity of Terriglobia in tundra soils.Environ Microbiol. 2024 May;26(5):e16640. doi: 10.1111/1462-2920.16640. Environ Microbiol. 2024. PMID: 38775217
-
Mucilaginibacter frigoritolerans sp. nov., Mucilaginibacter lappiensis sp. nov. and Mucilaginibacter mallensis sp. nov., isolated from soil and lichen samples.Int J Syst Evol Microbiol. 2010 Dec;60(Pt 12):2849-2856. doi: 10.1099/ijs.0.019364-0. Epub 2010 Jan 15. Int J Syst Evol Microbiol. 2010. PMID: 20081012
-
Mucilaginibacter paludis gen. nov., sp. nov. and Mucilaginibacter gracilis sp. nov., pectin-, xylan- and laminarin-degrading members of the family Sphingobacteriaceae from acidic Sphagnum peat bog.Int J Syst Evol Microbiol. 2007 Oct;57(Pt 10):2349-2354. doi: 10.1099/ijs.0.65100-0. Int J Syst Evol Microbiol. 2007. PMID: 17911309
-
Mucilaginibacter antarcticus sp. nov., isolated from tundra soil.Int J Syst Evol Microbiol. 2016 Dec;66(12):5140-5144. doi: 10.1099/ijsem.0.001486. Epub 2016 Sep 8. Int J Syst Evol Microbiol. 2016. PMID: 27613602
-
Effects on the function of Arctic ecosystems in the short- and long-term perspectives.Ambio. 2004 Nov;33(7):448-58. doi: 10.1579/0044-7447-33.7.448. Ambio. 2004. PMID: 15573572 Review.
References
-
- Bradshaw CJA, Warkentin IG. Global estimates of boreal forest carbon stocks and flux. Glob Planet Chang 2015;128:24–30. 10.1016/j.gloplacha.2015.02.004 - DOI
-
- Kirchhoff L, Gavazov K, Blume-Werry G et al. Microbial community composition unaffected by mycorrhizal plant removal in sub-arctic tundra. Fungal Ecol 2024;69:101342. 10.1016/j.funeco.2024.101342 - DOI
LinkOut - more resources
Full Text Sources
Research Materials

