Proteomics Analysis of the TDP-43 Interactome in Cellular Models of ALS Pathogenesis
- PMID: 40365763
- PMCID: PMC12076276
- DOI: 10.1111/jnc.70079
Proteomics Analysis of the TDP-43 Interactome in Cellular Models of ALS Pathogenesis
Abstract
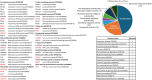
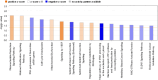
Cytoplasmic aggregation and nuclear depletion of TAR DNA-binding protein 43 (TDP-43) is a hallmark pathology of several neurodegenerative diseases including amyotrophic lateral sclerosis (ALS), frontotemporal lobar degeneration (FTLD) and limbic-predominant age-related TDP-43 encephalopathy (LATE). However, the protein interactome of TDP-43 remains incompletely defined. In this study, we aimed to identify putative TDP-43 protein partners within the nucleus and the cytoplasm and with different disease models of TDP-43 by comparing TDP-43 interaction partners in three different cell lines. We verified the levels of interaction of protein partners under stress conditions as well as after introducing TDP-43 variants containing ALS missense mutations (G294V and A315T). Overall, we identified 58 putative wild-type TDP-43 interactors, including novel binding partners responsible for RNA metabolism and splicing. Oxidative stress exposure broadly led to changes in TDP-43WT interactions with proteins involved in mRNA metabolism, suggesting a dysregulation of the transcriptional machinery early in disease. Conversely, although G294V and A315T mutations are both located in the C-terminal domain of TDP-43, both mutants presented different interactome profiles with most interaction partners involved in translational and transcriptional machinery. Overall, by correlating different cell lines and disease-simulating interventions, we provide a list of high-confidence TDP-43 interaction partners, including novel and previously reported proteins. Understanding pathological changes to TDP-43 and its specific interaction partners in different models of stress is critical to better understand TDP-43 proteinopathies and provide novel potential therapeutic targets and biomarkers.
Keywords: ALS; APEX; MND; TDP‐43; interactomics; proteomics.
© 2025 The Author(s). Journal of Neurochemistry published by John Wiley & Sons Ltd on behalf of International Society for Neurochemistry.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures





References
-
- Ansar, M. , Ullah F., Paracha S. A., et al. 2019. “Bi‐Allelic Variants in DYNC1I2 Cause Syndromic Microcephaly With Intellectual Disability, Cerebral Malformations, and Dysmorphic Facial Features.” American Journal of Human Genetics 104, no. 6: 1073–1087. 10.1016/j.ajhg.2019.04.002. - DOI - PMC - PubMed
-
- Arai, T. , Hasegawa M., Akiyama H., et al. 2006. “TDP‐43 Is a Component of Ubiquitin‐Positive Tau‐Negative Inclusions in Frontotemporal Lobar Degeneration and Amyotrophic Lateral Sclerosis.” Biochemical and Biophysical Research Communications 351, no. 3: 602–611. 10.1016/j.bbrc.2006.10.093. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

