Correlating the oral swab microbial community with milk production metrics in Holstein dairy cows
- PMID: 40366128
- PMCID: PMC12188706
- DOI: 10.1128/msphere.00167-25
Correlating the oral swab microbial community with milk production metrics in Holstein dairy cows
Abstract
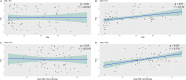
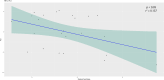
Oral swabs of dairy cows have been suggested as a proxy for direct ruminal sampling, and this approach can identify the presence of up to 70% of the rumen microbial community. Here, we further extend the utility of this approach by correlating the bacterial community of swabs collected from 226 dairy cows on a research farm in Wisconsin, USA, with average milk yield and days in milk, two phenotypes previously associated with differences in the ruminal microbiome. We then obtained milk production efficiency data for a subset of these animals (gross feed efficiency [GFE] and residual feed intake [RFI]) and correlated these metrics against their associated microbial data. We found that when using the oral swabs, we could identify correlations between bacterial genera and days in milk (P < 0.05). We further show that the ruminal microbiota was associated with average milk yield and days in milk for animals in their first lactation. Differential abundance testing identified amplicon sequence variants (ASVs) associated with these metrics (P < 0.05). Our comparison of bacterial communities between high and low efficiency groups, as determined by GFE and RFI, identified a significant difference in Shannon's diversity in second lactation cows (P < 0.05). We also found that RFI was significantly correlated with the bacterial community in second lactation animals (P < 0.05). Differential abundance analysis identified multiple oral- and rumen-associated ASVs correlated with GFE and RFI (P < 0.05). This study further establishes the utility of oral swabs as a ruminal proxy.IMPORTANCEImproving milk production efficiency is a key goal in the dairy industry and is traditionally pursued through genetic selection, diet optimization, and herd management practices. The ruminal microbiome, essential for digesting feed, has been linked to milk production efficiency, suggesting that microbiome modulation could improve efficiency. However, the integration of rumen microbiology into current management practices is hampered by the difficulty of large-scale rumen sampling, as proxies like fecal samples do not accurately reflect the ruminal microbiota. Traditional methods, like cannulation and stomach tubing, are labor-intensive and impractical for extensive sampling. Our research demonstrates the potential use of oral swabs as a scalable, effective method for characterizing the microbiome and its associations with milk production metrics, recapitulating established associations obtained through traditional ruminal sampling methods.
Keywords: milk production efficiency; milk yield; next-generation sequencing; oral community; oral swab; rumen community.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
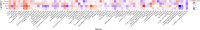
References
-
- Soest PJ. 1994. Nutritional ecology of the ruminant. Cornell University Press.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

