Dual oxic-anoxic co-culture enables direct study of anaerobe-host interactions at the airway epithelial interface
- PMID: 40366160
- PMCID: PMC12077211
- DOI: 10.1128/mbio.01338-24
Dual oxic-anoxic co-culture enables direct study of anaerobe-host interactions at the airway epithelial interface
Abstract
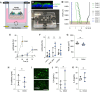
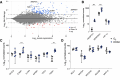
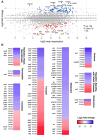
Strict and facultative anaerobic bacteria are widely associated with both acute and chronic airway diseases. However, their potential role(s) in disease pathophysiology remains poorly understood due to inherent limitations of existing laboratory models and conflicting oxygen demands between anaerobes and host cells. To address these limitations, here, we describe a dual oxic-anoxic culture (DOAC) approach that maintains an oxygen-limited microenvironment at the apical epithelial interface while host cells are oxygenated basolaterally. This platform enables epithelial-anaerobe co-culture for ~48 h, and we demonstrate its utility by evaluating reciprocal interactions between the oxygen-sensitive anaerobic bacterium, Fusobacterium nucleatum, and oxygen-demanding airway epithelial cells at the transcriptional level. Using bulk RNAseq, we demonstrate that epithelial colonization results in altered gene expression by F. nucleatum, highlighted by the differential expression of genes associated with virulence, ethanolamine and lysine metabolism, metal uptake, and other transport processes. We also combine DOAC with single-cell RNA sequencing to reveal a cell type-specific transcriptional response of the airway epithelium to F. nucleatum infection, including the increased expression of inflammatory marker genes and cancer-associated pathways. Together, these data illustrate the versatility of DOAC while revealing new insights into anaerobe-host interactions and their mechanistic contributions to airway disease pathophysiology.IMPORTANCEConflicting oxygen demands between anaerobes and host cells present a significant barrier to in vitro modeling of how these cell types interact. To this end, the significance of our dual oxic-anoxic culture (DOAC) approach lies in its ability to maintain anaerobe and epithelial viability during co-culture, paving the way for new insights into the role(s) of anaerobic microbiota in disease. We use DOAC to interrogate reciprocal interactions between the airway epithelium and Fusobacterium nucleatum-an anaerobic commensal with pathogenic potential. Given its link to a range of diseases, from localized infections to various cancers, these data showing how F. nucleatum bacterium re-shapes its metabolism and virulence upon epithelial colonization provide new mechanistic insight into F. nucleatum physiology and how the host responds. We use F. nucleatum as our model, but the DOAC platform motivates additional studies of the gut, lung, and oral cavity, where host-anaerobe interactions and the underlying mechanisms of pathogenesis are poorly understood.
Keywords: Fusobacterium nucleatum; RNAseq; airway epithelium; single-cell RNA sequencing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Segal LN, Alekseyenko AV, Clemente JC, Kulkarni R, Wu B, Gao Z, Chen H, Berger KI, Goldring RM, Rom WN, Blaser MJ, Weiden MD. 2013. Enrichment of lung microbiome with supraglottic taxa is associated with increased pulmonary inflammation. Microbiome 1:19. doi:10.1186/2049-2618-1-19 - DOI - PMC - PubMed
-
- Tunney MM, Field TR, Moriarty TF, Patrick S, Doering G, Muhlebach MS, Wolfgang MC, Boucher R, Gilpin DF, McDowell A, Elborn JS. 2008. Detection of anaerobic bacteria in high numbers in sputum from patients with cystic fibrosis. Am J Respir Crit Care Med 177:995–1001. doi:10.1164/rccm.200708-1151OC - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

