ANP32 proteins from ticks and vertebrates are key host factors for replication of Bourbon virus across species
- PMID: 40366164
- PMCID: PMC12172476
- DOI: 10.1128/jvi.00522-25
ANP32 proteins from ticks and vertebrates are key host factors for replication of Bourbon virus across species
Abstract
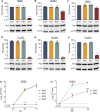
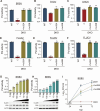
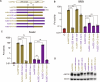
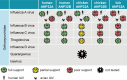
Bourbon virus (BRBV) is a tick-borne virus in the genus Thogotovirus in the Orthomyxoviridae family. BRBV was initially identified as the presumptive causative agent of a fatal human infection in 2014 and has since been identified in ticks in the Midwest, Northeast, and Southern United States, with occasional spillovers into humans. However, little is known about how virus-host interactions impact their large host range. Here, we show that BRBV polymerase activity in human cells is completely dependent on cellular ANP32 proteins. BRBV polymerase activity was completely lost in cells lacking ANP32A and ANP32B, resulting in failed infections. BRBV polymerase activity was restored in the presence of ANP32 proteins from diverse hosts. Dhori virus and Thogoto virus, other related Thogotovirus members, retained high activity in the absence of ANP32 proteins, showing reduced dependence on these host factors. Interaction studies revealed that the BRBV polymerase trimer binds human ANP32A or ANP32B. Genetic analysis revealed that tick vectors for BRBV encode a single ANP32 locus corresponding to ANP32A. Tick ANP32A produces multiple protein variants through alternative splicing and start-site selection, all of which enhance polymerase activity for Thogotoviruses. Unexpectedly, the BRBV polymerase was highly sensitive to changes at the N-terminus of ANP32, while it was insensitive to changes in the body of ANP32 that restrict the activity of influenza virus polymerases. Thus, ANP32A is a deeply conserved pro-viral cofactor, and Thogotoviruses show remarkable plasticity utilizing ANP32 homologs from different hosts separated by almost 1 billion years of evolution.IMPORTANCEViral polymerases rely on cellular cofactors to support efficient transcription of viral genes and replication of the viral genome. The RNA-dependent RNA polymerase of influenza virus, an orthomyxovirus, requires the cellular ANP32A or ANP32B proteins for genome replication. However, little is known about whether ANP32 proteins are required by other orthomyxovirus family members, like the tick-borne thogotoviruses. We show that thogotoviruses use ANP32 proteins from diverse hosts to enhance polymerase activity, including that encoded by the single ANP32A gene found in ticks. However, thogotovirus polymerase showed varying levels of dependence on ANP32 proteins, with some polymerases functioning at near full activity even in the absence of ANP32 proteins. Thus, ANP32 proteins are deeply conserved viral cofactors, with each virus displaying distinct patterns of ANP32 usage and requirements for function.
Keywords: Bourbon virus; anp32; host range; influenza virus; orthomyxovirus; thogotovirus; tick; virus:host interactions.
Conflict of interest statement
The Boon laboratory has received funding support unrelated to this work in sponsored research agreements from AI Therapeutics, GreenLight Biosciences Inc., Nano Targeting & Therapy Biopharma, and AbbVie.
Figures


References
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

