Assessing the sequencing success and analytical specificity of a targeted amplicon deep sequencing workflow for genotyping the foodborne parasite Cyclospora
- PMID: 40366167
- PMCID: PMC12153321
- DOI: 10.1128/jcm.01811-24
Assessing the sequencing success and analytical specificity of a targeted amplicon deep sequencing workflow for genotyping the foodborne parasite Cyclospora
Abstract
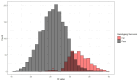
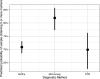
Epidemiological investigations of the foodborne parasitic illness cyclosporiasis can be aided by molecular techniques that enable the identification of genetically related clusters of Cyclospora isolates. At the Centers for Disease Control and Prevention (CDC), routine Cyclospora genotyping for the purpose of informing epidemiological outbreak investigations has occurred since 2018 using clinical stool specimens from case patients diagnosed with cyclosporiasis. This approach involves targeted amplicon deep sequencing of eight genotyping markers, followed by bioinformatic processing through a custom clustering algorithm. However, not all stool specimens submitted to the CDC for genotyping successfully amplify for at least five of the eight genotyping markers, the minimum required to be bioinformatically processed through the clustering algorithm. In this study, we utilized information from clinical stool specimens sent to the CDC from the years 2019 to 2023 to assess if the type of preservative, the age of the specimen, or the method used to diagnose the patient influenced the probability of successfully genotyping parasites from a fecal specimen. Additionally, we assessed the analytical specificity of the Cyclospora genotyping workflow by analyzing samples positive for other intestinal parasites, including closely related non-human infecting Cyclospora species and other coccidia. We found that stool specimens stored in preservatives had a greater likelihood of sequencing success over time relative to specimens without preservatives or those stored in non-nutritive transport media. Additionally, stool specimens from case patients diagnosed via microscopy-based methods were more likely to yield DNA of sufficient quality and quantity for genotyping compared to PCR or multiplex panels. Lastly, we determined that the genotyping workflow has an analytical specificity of 100%, as no non-human-infecting Cyclospora or other parasites yielded sequence data at >1 of the genotyping markers. This knowledge will help strengthen the quality of Cyclospora genotyping data produced in the future, improving the utility of this data for supporting epidemiological investigations.IMPORTANCEDetermining the genetic relatedness among parasites causing foodborne illness, such as Cyclospora, is a valuable tool to complement outbreak investigations. However, this molecular genotyping approach is limited by the quality and quantity of genetic data obtained from the samples being investigated. In this study, we demonstrate that the storage conditions of clinical stool specimens are correlated to the quality of sequence data produced for Cyclospora genotyping. Our insights can be used to guide storage recommendations for stool specimens, which can improve the quality of foodborne illness outbreak investigations conducted in the future. Additionally, we showed that the current Cyclospora genotyping tool used by the Centers for Disease Control (CDC) is highly specific to human-infecting Cyclospora parasites; this valuable information indicates that the CDC's Cyclospora investigations are not negatively impacted by false-positive detections.
Keywords: cyclosporiasis; foodborne illness; molecular epidemiology; parasitology.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- CDC . 2024. Multistate foodborne outbreak notices. Available from: https://www.cdc.gov/foodborne-outbreaks/active-investigations/all-foodbo.... Retrieved 20 Sep 2024.
-
- Administration UFaD . 2020. Del Monte Fresh Produce N.A., Inc. Voluntarily Recalls Limited Quantity of Vegetable Trays in A Multistate Outbreak of Cyclospora Illnesses in Select Retailers in Illinois, Indiana, Iowa, Michigan, Minnesota and Wisconsin, Because of Possible Health Risk. Available from: https://www.fda.gov/safety/recalls-market-withdrawals-safety-alerts/del-.... Retrieved 20 Sep 2024.
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

