A single amino acid mutation in VP1 of coxsackievirus A6 determining efficiency of VP0 cleavage and proliferation
- PMID: 40366174
- PMCID: PMC12180516
- DOI: 10.1128/jvi.00128-25
A single amino acid mutation in VP1 of coxsackievirus A6 determining efficiency of VP0 cleavage and proliferation
Abstract
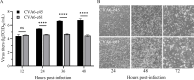
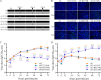
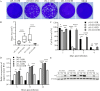
Coxsackievirus A6 (CV-A6) has emerged as a major pathogen associated with hand, foot, and mouth disease (HFMD), capable of infecting both children and adults. However, currently, there is no effective vaccine to prevent HFMD caused by non-EV-A71 enteroviruses. In this study, a pair of CV-A6 strains was selected from a rhabdomyosarcoma (RD)-isolated and Vero-adapted stock with a difference of 7 nucleotides in their genomes, resulting in three amino acid mutations in the structural proteins. Distinct differences in propagation, virulence in cells, and plaque size were observed. A series of single-site mutants was constructed, and a single mutation in VP1-143 was mapped to associate with phenotype changes. The mutation from glycine to arginine at VP1-143 dramatically increased infectivity but decreased virulence, growth rate, and plaque size. Furthermore, the experiments using both purified whole virus and full particle (FP) demonstrated that glycine-to-arginine mutation increased VP0 cleavage efficiency because of decreased VP0/VP2 ratio. The decrease in VP0 cleavage efficiency led to the accumulation of non-infectious provirion. The efficiency of virus transmission between cells determined the rates of viral RNA (vRNA) and protein synthesis and was related to fast-slow growth and virulence phenotypes. In addition, the data indicated that the mutation did not affect the encapsidation of the genomic RNA, and the ratio of empty and full particles was unchanged. The results are important for understanding the mechanism of VP0 cleavage regulation and are relevant to developing vaccines and therapeutic reagents against CV-A6 infection and diseases.
Importance: CV-A6 is a major pathogen in the context of HFMD. The cost of treatment and hospitalization of children with HFMD may have a considerable financial impact on the families of patients. CV-A6 is a member of picornaviruses and forms infectious virion through maturation cleavage of VP0 into VP4 and VP2. Although it is well accepted that the autocatalytic process involves viral RNA, the detailed mechanism remains unclear. In this study, residues in VP1-143 were demonstrated to regulate the efficiency of VP0 cleavage and affect the ratio of provirion and virion. Glycine-to-arginine mutation was tolerant, not abolished, but affected the efficiency of VP0 cleavage. The results support a theory that residue mutations on a structural protein of a serotype/genotype within enteroviruses, not well-conserved across picornaviruses and far away from the VP0 cleavage site on the outside surface, regulate the efficiency of VP0 cleavage and render phenotypically different strains.
Keywords: CV-A6; VP0 cleavage regulation; proliferation; virulence.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Broccolo F, Drago F, Ciccarese G, Genoni A, Puggioni A, Rosa GM, Parodi A, Manukyan H, Laassri M, Chumakov K, Toniolo A. 2019. Severe atypical hand-foot-and-mouth disease in adults due to coxsackievirus A6: clinical presentation and phylogenesis of CV-A6 strains. J Clin Virol 110:1–6. doi: 10.1016/j.jcv.2018.11.003 - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

