Clicking viruses-with chemistry toward mechanisms in infection
- PMID: 40366176
- PMCID: PMC12172498
- DOI: 10.1128/jvi.00471-25
Clicking viruses-with chemistry toward mechanisms in infection
Abstract
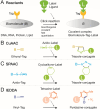
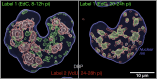
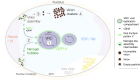
Viruses subvert cells and evade host defense. They emerge unpredictably and threaten humans and livestock through their genetic and phenotypic diversity. Despite more than 100 years since the discovery of viruses, the molecular underpinnings of virus infections are incompletely understood. The introduction of new methodologies into the field, such as that of click chemistry some 10 years ago, keeps uncovering new facets of viruses. Click chemistry uses bio-orthogonal reactions on chemical probes and couples nucleic acids, proteins, and lipids with tractable labels, such as fluorophores for single-cell and single-molecule imaging, or biotin for biochemical profiling of infections. Its applications in single cells often achieve single-molecule resolution and provide important insights into the widely known phenomenon of cell-to-cell infection variability. This review describes click chemistry advances to unravel infection mechanisms of a select set of enveloped and nonenveloped DNA and RNA viruses, including adenovirus, herpesvirus, and human immunodeficiency virus. It highlights recent click chemistry breakthroughs with viral DNA, viral RNA, protein, as well as host-derived lipid functions in both live and chemically fixed cells. It discusses new insights on specific processes including virus entry, uncoating, transcription, replication, packaging, and assembly and provides a perspective for click chemistry to explore viral cell biology, infection variability, and genome organization in the particle.
Keywords: DNA; RNA; adenovirus; click chemistry; herpesvirus; human immunodeficiency virus (HIV); lipid; protein; virus entry; virus genome packaging and assembly.
Conflict of interest statement
The author declares no conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

