An obligate symbiont of Haematomyzus elephantis with a strongly reduced genome resembles symbiotic bacteria in sucking lice
- PMID: 40366182
- PMCID: PMC12175528
- DOI: 10.1128/aem.00220-25
An obligate symbiont of Haematomyzus elephantis with a strongly reduced genome resembles symbiotic bacteria in sucking lice
Abstract
The parvorder Rhynchophthirina with a single genus Haematomyzus is a small group of ectoparasites of unclear phylogenetic position, related to sucking and chewing lice. Previous screening based on the 16S rRNA gene indicated that Haematomyzus harbors a symbiotic bacterium whose DNA exhibits a strong shift in nucleotide composition typical of obligate mutualistic symbionts in insects. Within Phthiraptera, the smallest known genomes are found in the symbionts associated with sucking lice, which feed exclusively on mammal blood, compared to the generally larger genomes of the symbionts inhabiting chewing lice, which feed on skin derivatives. In this study, we investigate the genome characteristics of the symbiont associated with Haematomyzus elephantis. We sequenced and assembled the H. elephantis metagenome, extracted a genome draft of its symbiotic bacterium, and showed that the symbiont has a significantly reduced genome, which is with 0.39 Mbp the smallest genome among the symbionts known from Phthiraptera. Multigenic phylogenetic analysis places the symbiont into one of three clusters composed of long-branched symbionts from other insects. More specifically, it clusters together with symbionts from several other sucking lice and also with Wigglesworthia glossinidia, an obligate symbiont of tsetse flies. Consistent with the dramatic reduction of its genome, the H. elephantis symbiont lost many metabolic capacities. However, it retained functional pathways for four B vitamins, a trait typical for symbionts in blood-feeding insects. Considering genomic, metabolic, and phylogenetic characteristics, the new symbiont closely resembles those known from several sucking lice rather than chewing lice.IMPORTANCERhynchophthirina is a unique small group of permanent ectoparasites that is closely related to both sucking and chewing lice. These two groups of lice differ in their morphology, ecology, and feeding strategies. As a consequence of their different dietary sources, i.e., mammals' blood vs vertebrate skin derivatives, they also exhibit distinct patterns of symbiosis with obligate bacterial symbionts. While Rhynchophthirina shares certain traits with sucking and chewing lice, the nature of its obligate symbiotic bacterium and its metabolic role is not known. In this study, we assemble the genome of symbiotic bacterium from Haematomyzus elephantis (Rhynchophthirina), demonstrating its close similarity and phylogenetic proximity to several symbionts of sucking lice. The genome is highly reduced (representing the smallest genome among louse-associated symbionts) and exhibits a significant loss of metabolic pathways. However, similar to other sucking louse symbionts, it retains essential pathways for the synthesis of several B vitamins.
Keywords: endosymbionts; genomics; lice.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
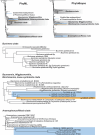
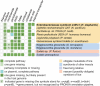
References
-
- de Moya RS, Yoshizawa K, Walden KKO, Sweet AD, Dietrich CH, Kevin P J. 2021. Phylogenomics of parasitic and nonparasitic lice (Insecta: Psocodea): combining sequence data and exploring compositional bias solutions in next generation data sets. Syst Biol 70:719–738. doi: 10.1093/sysbio/syaa075 - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

