A Holistic Investigation of Arabidopsis Proteomes Altered in Chloroplast Biogenesis and Retrograde Signalling Identifies PsbO as a Key Regulator of Chloroplast Quality Control
- PMID: 40366233
- PMCID: PMC12223713
- DOI: 10.1111/pce.15611
A Holistic Investigation of Arabidopsis Proteomes Altered in Chloroplast Biogenesis and Retrograde Signalling Identifies PsbO as a Key Regulator of Chloroplast Quality Control
Abstract
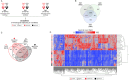
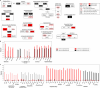
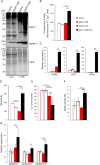
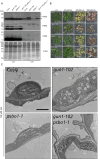
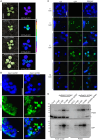
Communication between the diverse compartments of plant cells relies on an intricate network of molecular interactions that orchestrate organellar development and adaptation to environmental conditions. Plastid-to-nucleus signalling pathways play a key role in relaying information from developing, mature, and damaged or disintegrating chloroplasts to the nucleus, which serves to coordinate gene expression between the two genomes. To shed light on these mechanisms, we performed a comprehensive analysis of the response of the Arabidopsis thaliana proteomes to perturbation of chloroplast biogenesis by the antibiotic lincomycin (Lin) in the absence of GENOMES UNCOUPLED 1 (GUN1), a key player in plastid-to-nucleus signalling. The topological analysis of protein-protein interactions (PPIs) and co-expression networks enabled the identification of protein hubs in each genotype and condition tested, and highlighted whole-cell adaptive responses to the disruption of chloroplast biogenesis. Our findings reveal a novel role for PsbO, a subunit of the oxygen-evolving complex (OEC), which behaves as an atypical photosynthetic protein upon inhibition of plastid protein synthesis. Notably, and unlike all other subunits of the thylakoid electron transport chain, PsbO accumulates in non-photosynthetic plastids, and is crucial for the breakdown of damaged chloroplasts.
Keywords: chloroplast biology; chloroplast degradation; intracellular signalling; oxidative stress; proteome; proteomics.
© 2025 The Author(s). Plant, Cell & Environment published by John Wiley & Sons Ltd.
Figures



References
-
- Allahverdiyeva, Y. , Suorsa M., Rossi F., et al. 2013. “ Arabidopsis Plants Lacking PsbQ and PsbR Subunits of the Oxygen‐Evolving Complex Show Altered PSII Super‐Complex Organization and Short‐Term Adaptive Mechanisms.” Plant Journal 75: 671–684. - PubMed
-
- Anjum, N. A. , Sharma P., Gill S. S., et al. 2016. “Catalase and Ascorbate Peroxidase—Representative H2O2‐detoxifying Heme Enzymes in Plants.” Environmental Science and Pollution Research 23: 19002–19029. - PubMed
-
- Assenov, Y. , Ramírez F., Schelhorn S.‐E., Lengauer T., and Albrecht M.. 2008. “Computing Topological Parameters of Biological Networks.” Bioinformatics 24: 282–284. - PubMed

