Identification of a Biomarker Panel in Extracellular Vesicles Derived From Non-Small Cell Lung Cancer (NSCLC) Through Proteomic Analysis and Machine Learning
- PMID: 40366616
- PMCID: PMC12077270
- DOI: 10.1002/jev2.70078
Identification of a Biomarker Panel in Extracellular Vesicles Derived From Non-Small Cell Lung Cancer (NSCLC) Through Proteomic Analysis and Machine Learning
Abstract
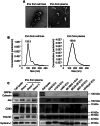
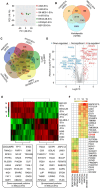
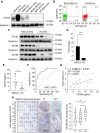
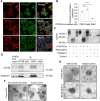
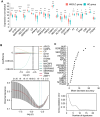
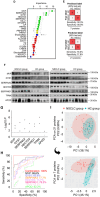
Antigen fingerprint profiling of tumour-derived extracellular vesicles (TDEVs) in the body fluids is a promising strategy for identifying tumour biomarkers. In this study, proteomic and immunological assays reveal significantly higher CD155 levels in plasma extracellular vesicles (EVs) from patients with non-small cell lung cancer (NSCLC) than from healthy individuals. Utilizing CD155 as a bait protein on the EV membrane, CD155+ TDEVs are enriched from NSCLC patient plasma EVs. In the discovery cohort, 281 differentially expressed proteins are identified in TDEVs of the NSCLC group compared with the healthy control group. In the verification cohort, 49 candidate biomarkers are detected using targeted proteomic analysis. Of these, a biomarker panel of seven frequently and stably detected proteins-MVP, GYS1, SERPINA3, HECTD3, SERPING1, TPM4, and APOD-demonstrates good diagnostic performance, achieving an area under the curve (AUC) of 1.0 with 100% sensitivity and specificity in receiver operating characteristic (ROC) curve analysis, and 92.3% sensitivity and 88.9% specificity in confusion matrix analysis. Western blotting results confirm upregulation trends for MVP, GYS1, SERPINA3, HECTD3, SERPING1 and APOD, and TPM4 is downregulated in EVs of NSCLC patients compared with healthy individuals. These findings highlight the potential of this biomarker panel for the clinical diagnosis of NSCLC.
Keywords: NSCLC; biomarkers; diagnosis; extracellular vesicles; proteomics.
© 2025 The Author(s). Journal of Extracellular Vesicles published by Wiley Periodicals LLC on behalf of International Society for Extracellular Vesicles.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

