Thymic dendritic cell-derived IL-27p28 promotes the establishment of functional bias against IFN-γ production in newly generated CD4+ T cells through STAT1-related epigenetic mechanisms
- PMID: 40366856
- PMCID: PMC12077877
- DOI: 10.7554/eLife.96868
Thymic dendritic cell-derived IL-27p28 promotes the establishment of functional bias against IFN-γ production in newly generated CD4+ T cells through STAT1-related epigenetic mechanisms
Abstract
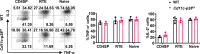
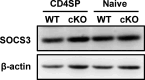
The newly generated CD4 single-positive (SP) T lymphocytes are featured by enhanced IL-4 but repressed IFN-γ production. The mechanisms underlying this functional bias remain elusive. Previous studies have reported that CD4+ T cells from mice harboring dendritic cell (DC)-specific deletion of IL-27p28 display an increased capacity of IFN-γ production upon TCR stimulation. Here, we demonstrated that similarly altered functionality occurred in CD4SP thymocytes, recent thymic emigrants (RTEs), as well as naive T cells from either Cd11c-p28f/f mice or mice deficient in the α subunit of IL-27 receptor. Therefore, DC-derived IL-27p28-triggered, IL-27Rα-mediated signal is critically involved in the establishment of functional bias against IFN-γ production during their development in the thymus. Epigenetic analyses indicated reduced DNA methylation of the Ifng locus and increased trimethylation of H3K4 at both Ifng and Tbx21 loci in CD4SP thymocytes from Cd11c-p28f/f mice. Transcriptome profiling demonstrated that Il27p28 ablation resulted in the coordinated up-regulation of STAT1-activated genes. Concurrently, STAT1 was found to be constitutively activated. Moreover, we observed increased accumulation of STAT1 at the Ifng and Tbx21 loci and a strong correlation between STAT1 binding and H3K4me3 modification of these loci. Of note, Il27p28 deficiency exacerbated the autoimmune phenotype of Aire-/- mice. Collectively, this study reveals a novel mechanism underlying the functional bias of newly generated CD4+ T cells and the potential relevance of such a bias in autoimmunity.
Keywords: IFN-γ; IL-27p28; Th2 bias; autoimmune disease; immunology; inflammation; mouse; single positive thymocyte.
© 2024, Zhang et al.
Conflict of interest statement
JZ, HT, HW, XP, RJ, YZ No competing interests declared
Figures












Update of
- doi: 10.1101/2024.03.02.583138
- doi: 10.7554/eLife.96868.1
- doi: 10.7554/eLife.96868.2
References
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

