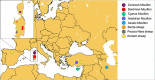
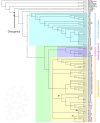
Revised phylogeny of mouflon based on expanded sampling of mitogenomes
- PMID: 40367058
- PMCID: PMC12077669
- DOI: 10.1371/journal.pone.0323354
Revised phylogeny of mouflon based on expanded sampling of mitogenomes
Abstract
Mouflons are flagship species of the Mediterranean islands where they persist. Once thought to be the remnants of a European wild sheep population, archaeology suggests they were introduced by humans to the islands of Cyprus in the Early Neolithic (~10,000 years ago) and later to Corsica and Sardinia. Their status as truly wild animals remains a subject of debate. To investigate the phylogenetic relationship between these island populations and other domestic and wild sheep from the Mediterranean region, we sequenced 50 mitogenomes of mouflons from Sardinia and Corsica, and modern and ancient Sardinian domestic sheep. A total of 68 additional publicly available mitogenomes were included in the comparative analysis and used to reconstruct the phylogeny of sheep and its closest wild relative, the mouflon (Ovis gmelini). Our study analyzed the evolutionary relationships within the C-E-X and haplogroup B clusters, showing that: a) Cyprus mouflons are more related to Anatolian and Iranian mouflons belonging to the wild haplogroup X, which seems to be basal to the domestic C and E haplogroups; b) Corsican and Sardinian mouflon arise from basal lineages associated with the early European expansion of domestic sheep. These results highlight the phylogenetic distinctiveness of the mouflon populations from the Mediterranean islands, suggesting a revision of their systematic classification and an update of the nomenclature for Sardinian and Corsican mouflons from the current status of subspecies of domestic sheep (Ovis aries musimon) to subspecies of their wild relatives (Ovis gmelini musimon) which would facilitate conservation efforts.
Copyright: © 2025 Mereu et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
The First Mitogenome of the Cyprus Mouflon (Ovis gmelini ophion): New Insights into the Phylogeny of the Genus Ovis.PLoS One. 2015 Dec 4;10(12):e0144257. doi: 10.1371/journal.pone.0144257. eCollection 2015. PLoS One. 2015. PMID: 26636977 Free PMC article.
-
Genomic signatures of adaptive introgression from European mouflon into domestic sheep.Sci Rep. 2017 Aug 8;7(1):7623. doi: 10.1038/s41598-017-07382-7. Sci Rep. 2017. PMID: 28790322 Free PMC article.
-
Genomic analyses of Asiatic Mouflon in Iran provide insights into the domestication and evolution of sheep.Genet Sel Evol. 2025 Jun 13;57(1):31. doi: 10.1186/s12711-025-00978-y. Genet Sel Evol. 2025. PMID: 40514664 Free PMC article.
-
Population Genomic History of the Endangered Anatolian and Cyprian Mouflons in Relation to Worldwide Wild, Feral, and Domestic Sheep Lineages.Genome Biol Evol. 2024 May 2;16(5):evae090. doi: 10.1093/gbe/evae090. Genome Biol Evol. 2024. PMID: 38670119 Free PMC article.
-
Phenotype transition from wild mouflon to domestic sheep.Genet Sel Evol. 2024 Jan 2;56(1):1. doi: 10.1186/s12711-023-00871-6. Genet Sel Evol. 2024. PMID: 38166592 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources