Interleukin-4 modulates type I interferon to augment antitumor immunity
- PMID: 40367186
- PMCID: PMC12077506
- DOI: 10.1126/sciadv.adt3618
Interleukin-4 modulates type I interferon to augment antitumor immunity
Abstract
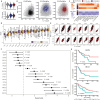
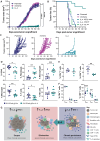
Despite advances in immunotherapy, metastatic melanoma remains a considerable therapeutic challenge due to the complexity of the tumor microenvironment. Intratumoral type I interferon (IFN-I) has long been associated with improved clinical outcomes. However, several IFN-I subtypes can also paradoxically promote tumor growth in some contexts. We investigated this further by engineering murine B16 melanoma cells to overexpress various IFN-I subtypes, where a spectrum of outcomes was observed. Characterization of these tumors by RNA sequencing revealed a tumor immune phenotype, where potent IFN-I signaling concomitant with diminished type 2 inflammation failed to confer durable tumor control. T cell-mediated rejection of these tumors was restored by introducing interleukin-4 (IL-4) into the tumor microenvironment, either through ectopic expression or in a preclinical adoptive T cell therapy model. Collectively, our findings highlight the IFN-I/IL-4 axis in promoting antitumor immunity, which could be harnessed to target and stratify solid tumors that are nonresponsive to frontline therapies.
Figures




References
-
- Duan Q., Zhang H., Zheng J., Zhang L., Turning cold into hot: Firing up the tumor microenvironment. Trends Cancer 6, 605–618 (2020). - PubMed
-
- Klein S., Schulte A., Arolt C., Tolkach Y., Reinhardt H. C., Buettner R., Quaas A., Intratumoral abundance of M2-macrophages is associated with unfavorable prognosis and markers of T-cell exhaustion in small cell lung cancer patients. Mod. Pathol. 36, 100272 (2023). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

