Top-Down and Middle-Down Mass Spectrometry of Antibodies
- PMID: 40368137
- PMCID: PMC12343474
- DOI: 10.1016/j.mcpro.2025.100989
Top-Down and Middle-Down Mass Spectrometry of Antibodies
Abstract
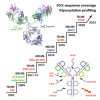
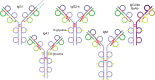
Therapeutic antibodies, primarily immunoglobulin G-based monoclonal antibodies, are developed to treat cancer, autoimmune disorders, and infectious diseases. Their large size, structural complexity, and heterogeneity pose significant analytical challenges, requiring advanced characterization techniques. This review traces the 30-year evolution of top-down (TD) and middle-down (MD) mass spectrometry (MS) for antibody analysis, beginning with their initial applications and highlighting key advances and challenges throughout this period. TD MS allows for the analysis of intact antibodies, and MD MS performs analysis of the antibody subunits, even in complex biological samples. Both approaches preserve critical quality attributes such as sequence integrity, post-translational modifications (PTMs), disulfide bonds, and glycosylation patterns. Key milestones in TD and MD MS of antibodies include the use of structure-specific enzymes for subunit generation, the implementation of high-resolution mass spectrometers, and the adoption of non-ergodic ion activation methods such as electron transfer dissociation (ETD), electron capture dissociation (ECD), ultraviolet photodissociation (UVPD), and matrix-assisted laser desorption/ionization in-source decay (MALDI-ISD). The combination of complementary dissociation methods and consecutive ion activation approaches has further enhanced TD/MD MS performance. The current TD MS record of antibody sequencing with terminal product ions is about 60% sequence coverage obtained using the activated ion-ETD approach on a high-resolution MS platform. Current MD MS analyses with about 95% sequence coverage were achieved using combinations of ion activation and dissociation techniques. The review explores TD and MD MS analysis of novel mAb modalities, including antibody-drug conjugates, bispecific antibodies, endogenous antibodies from biofluids, and immunoglobulin A and M-type classes.
Keywords: ADC; ETD; Orbitrap; antibody-drug conjugate; electron transfer dissociation; mAb; middle-down mass spectrometry; monoclonal antibody; proteoform; top-down mass spectrometry.
Copyright © 2025 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare the following competing financial interest(s): N.A.K is an employee of Spectrotech SAS and A. N. K., K. O. N., and Y. O. T. are employees of Spectroswiss Sarl. Both companies develop and commercialize FTMS data acquisition, processing, and analysis tools.
Figures




References
-
- Campuzano I.D.G., Sandoval W. Denaturing and native mass spectrometric analytics for biotherapeutic drug discovery research: historical, current, and future personal perspectives. J. Am. Soc. Mass Spectrom. 2021;32:1861–1885. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

