Macrophage-augmented intestinal organoids model virus-host interactions in enteric viral diseases and facilitate therapeutic development
- PMID: 40368896
- PMCID: PMC12078800
- DOI: 10.1038/s41467-025-59639-9
Macrophage-augmented intestinal organoids model virus-host interactions in enteric viral diseases and facilitate therapeutic development
Abstract
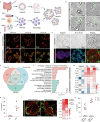
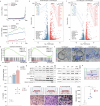
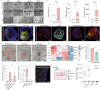
The pathogenesis of enteric viral infections is attributed to both viral replication and the resultant immune-inflammatory response. To recapitulate this complex pathophysiology, we engineer macrophage-augmented organoids (MaugOs) by integrating human macrophages into primary intestinal organoids. Echovirus 1, echovirus 6, rotavirus, seasonal coronavirus OC43 and SARS-CoV-2- known to directly invade the intestine- are used as disease modalities. We demonstrate that these viruses efficiently propagate in MaugOs and stimulate the host antiviral response. However, rotavirus, coronavirus OC43 and SARS-CoV-2, but not the two echoviruses, trigger inflammatory responses. Acetate, a microbial metabolite abundantly present in the intestine, potently inhibits virus-induced inflammatory responses in MaugOs, while differentially affecting viral replication in macrophages and organoids. Furthermore, we provide a proof-of-concept of combining antiviral agent with either anti-inflammatory regimen or acetate to simultaneously inhibit viral infection and inflammatory response in MaugOs. Collectively, these findings demonstrate that MaugOs are innovative tools for studying the complex virus-host interactions and advancing therapeutic development.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

