Molecular mechanism of thyroxine transport by monocarboxylate transporters
- PMID: 40368961
- PMCID: PMC12078798
- DOI: 10.1038/s41467-025-59751-w
Molecular mechanism of thyroxine transport by monocarboxylate transporters
Abstract
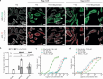
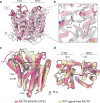
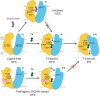
Thyroid hormones (the common name for prohormone thyroxine and the bioactive form triiodothyronine) control major developmental and metabolic processes. Release of thyroid hormones from the thyroid gland into the bloodstream and their transport into target cells is facilitated by plasma membrane transporters, including monocarboxylate transporter (MCT)8 and the highly homologous MCT10. However, the molecular mechanism underlying thyroid hormone transport is unknown. The relevance of such transporters is illustrated in patients with MCT8 deficiency, a severe neurodevelopmental and metabolic disorder. Using cryogenic-sample electron microscopy (cryo-EM), we determined the ligand-free and thyroxine-bound human MCT8 structures in the outward-facing state and the thyroxine-bound human MCT10 in the inward-facing state. Our structural analysis revealed a network of conserved gate residues involved in conformational changes upon thyroxine binding, triggering ligand release in the opposite compartment. We then determined the structure of a folded but inactive patient-derived MCT8 mutant, indicating a subtle conformational change which explains its reduced transport activity. Finally, we report a structure of MCT8 bound to its inhibitor silychristin, locked in the outward-facing state, revealing the molecular basis of its action and specificity. Taken together, this study advances mechanistic understanding of normal and disordered thyroid hormone transport.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures


References
-
- Friesema, E. C. H. et al. Identification of monocarboxylate transporter 8 as a specific thyroid hormone transporter. J. Biol. Chem.278, 40128–40135 (2003). - PubMed
-
- Groeneweg, S., van Geest, F. S., Peeters, R. P., Heuer, H. & Visser, W. E. Thyroid hormone transporters. Endocr. Rev.41, bnz008 (2020). - PubMed
-
- Bernal, J., Guadaño-Ferraz, A. & Morte, B. Thyroid hormone transporters–functions and clinical implications. Nat. Rev. Endocrinol.11, 406–417 (2015). - PubMed
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources

