Interferon-γ orchestrates leptomeningeal anti-tumour response
- PMID: 40369076
- PMCID: PMC12286854
- DOI: 10.1038/s41586-025-09012-z
Interferon-γ orchestrates leptomeningeal anti-tumour response
Abstract
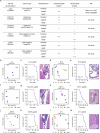
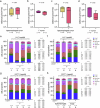
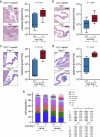
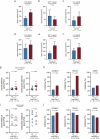
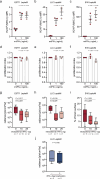
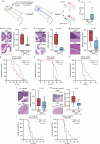
Metastasis to the cerebrospinal-fluid-filled leptomeninges, or leptomeningeal metastasis, represents a fatal complication of solid tumours1. Multimodal analyses of clinical specimens reveal substantial inflammatory infiltrate in leptomeningeal metastases with enrichment of IFNγ and resulting downstream signalling. Here, to investigate and overcome this futile anti-tumour response within the leptomeninges, we developed syngeneic lung cancer, breast cancer and melanoma leptomeningeal-metastasis mouse models. We show that transgenic host mice lacking IFNγ or its receptor fail to control the growth of leptomeningeal metastases growth. Leptomeningeal overexpression of Ifng through a targeted adeno-associated-virus-based system controls cancer cell growth independent of adaptive immunity. Using a suite of transgenic hosts, we demonstrate that leptomeningeal T cells generate IFNγ to actively recruit and activate peripheral myeloid cells, generating a diverse spectrum of dendritic cell subsets. Independent of antigen presentation, migratory CCR7+ dendritic cells orchestrate the influx, proliferation and cytotoxic action of natural killer cells to control cancer cell growth in the leptomeninges. This study identifies unique, leptomeninges-specific IFNγ signalling and suggests an immune-therapeutic approach against tumours within this space.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: J.R. and A.B. are listed as inventors on provisional patent applications based on this study, filed by MSKCC. A.B. holds an unpaid position on the scientific advisory board for Evren Scientific and is listed as an inventor on the following patents: 62/258,044, 10/413,522 and 63/052,139. D.P. is on the scientific advisory board of Insitro. The other authors declare no competing interests.
Figures













References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

