Synergistic effects of LCN2 and TWEAK on the progression of psoriasis
- PMID: 40369189
- PMCID: PMC12206918
- DOI: 10.1038/s41423-025-01292-9
Synergistic effects of LCN2 and TWEAK on the progression of psoriasis
Abstract
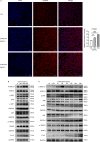
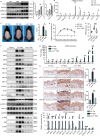
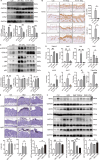
Lipocalin 2 (LCN2) and the TWEAK/Fn14 signaling pathways are pivotal in psoriasis, influencing epidermal development, inflammatory cell chemotaxis, and inflammatory factor release. Despite their significant roles, the intricate relationship between LCN2 and TWEAK/Fn14 pathways remains unclear. Our study revealed the correlation between the expression of TWEAK, LCN2, and Fn14 in psoriatic lesions. We found that TWEAK is expressed by keratinocytes and macrophages, while LCN2 is expressed by keratinocytes and neutrophils. Surface plasmon resonance experiments demonstrated binding between LCN2 and Fn14, which was further validated by co-immunoprecipitation and cellular co-localization via immunofluorescence. In vitro, LCN2 promoted macrophage differentiation and TWEAK secretion, enhanced TWEAK and Fn14 expression in keratinocytes, and activated the MAPK signaling pathway. TWEAK upregulated LCN2 expression in neutrophils but not in keratinocytes. Bulk RNA-seq analysis revealed a synergistic effect of LCN2 and TWEAK in promoting inflammatory cytokine expression in keratinocytes, with enhanced MAPK pathway activation in the presence of M5 cytokines. Lcn2 knockout reduced Fn14 expression in skin lesions and serum TWEAK levels of imiquimod-induced murine psoriasis model, while Fn14 knockout attenuated the epidermal hyperplasia-promoting effects of TWEAK and LCN2. Overexpression of Fn14 in keratinocytes led to higher TWEAK expression upon LCN2 stimulation, suggesting a self-reinforcing loop among TWEAK, LCN2, and Fn14. We propose that LCN2 synergizes with TWEAK through Fn14 to drive psoriasis pathogenesis.
Keywords: Fn14; Lipocalin 2; Psoriasis; TWEAK; synergistic.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

