BCR::ABL1 expression in chronic myeloid leukemia cells in low oxygen is regulated by glutamine via CD36-mediated fatty acid uptake
- PMID: 40369538
- PMCID: PMC12080266
- DOI: 10.1186/s12935-025-03805-y
BCR::ABL1 expression in chronic myeloid leukemia cells in low oxygen is regulated by glutamine via CD36-mediated fatty acid uptake
Abstract
Background: Chronic myeloid leukemia (CML) is influenced by microenvironmental nutrients, glucose (Glc), and glutamine (Gln) which regulate cell proliferation, viability, and the expression of the driver oncoprotein (BCR::ABL1).
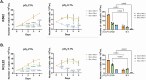
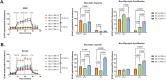
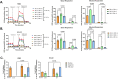
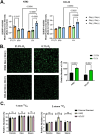
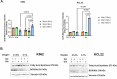
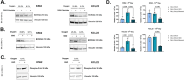
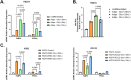
Results: Our study revealed that Glc, while partially supporting alone cell growth in normoxia, is essential in low oxygen conditions, whereas Gln is ineffective. Under low oxygen, Gln reduced oxidative respiratory activity while enhancing glycolysis. In these conditions, fatty acid (FA) metabolism becomes crucial, as evidenced by increased lipid droplets (LD) accumulation when Glc was absent. Gln, in particular, drives CD36-mediated FA uptake, suppressing the BCR::ABL1 oncoprotein and facilitating cell survival. By co-culturing leukemia cells with adipocytes, one of the main bone marrow (BM) cell components, we observed an enhanced FA release, suggesting a link between FA, microenvironmental BM cells, and the maintenance of leukemic stem cells (LSC).
Methods: K562 and KCL22 cell lines were subjected to Glc and/or Gln deprivation under hypoxic conditions (96 h at 0.1% O2). Metabolic profiling was conducted through the Seahorse XFe96 analyzer, and the contribution of L-Glutamine-13C5 to FA de novo synthesis was determined via GC/MS. Intracellular neutral LD were measured using BODIPY 493/503 in confocal microscopy and flow cytometry, with their presence and morphology further examined via transmission electron microscopy. BCR::ABL1 as well as several FA-related markers were evaluated via Western Blotting, whilst CD36 was determined through flow cytometry. LC2 assay was used for measuring leukemia stem cell potential by inhibiting FA uptake via the usage of the Sulfo-N-Succinimidyl Oleate, a CD36 inhibitor. qPCR was exploited to detect markers of FA secretion in CML-adipocytes co-culture together with Nile Red staining to assess free FA in the media.
Conclusions: These findings underscore the central role of FA in the regulation of the LSC compartment of CML, highlighting the importance of Gln in facilitating CML cell survival under restrictive metabolic conditions and preparing the cell population for expansion upon the release of these restrictions.
Keywords: BCR:ABL1; Chronic myeloid leukemia; Fatty acids; Hypoxia.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures

References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

